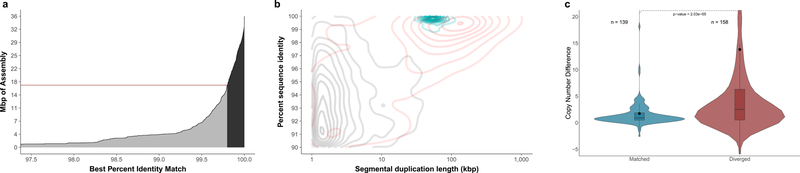
Figure 2. SDA results of the CHM1 human genome assembly.
a) A cumulative distribution of the SDA assemblies and their percent identity to their best match in the reference. There is 16.4 Mbp of diverged assembly (<99.8% identity, gray) and 18.8 Mbp that map to the reference at high identity (>99.8% identity, black). The number of assembly Mbp is calculated independently of a mapping to the reference, unlike in Table 1. b) Density plot of SDs plotted by length and percent identity. Black represents duplications resolved in the CHM1 assembly, red shows unresolved duplications in the CHM1 assembly, and blue represents paralogs assembled using SDA. Resolved SDA sequences are “content” resolved and not ordered within the genome, whereas SDs in the assembly must extend into unique sequence on both sides to be considered resolved. c) Copy number difference (CND) between CHM1 and the reference genome (CHM1 copy number – reference genome copy number) comparing n=139 SD regions that match (>99.8%) versus n=158 diverged SD regions (<99.8% identity). The mean CND of the matched sequence is 1.75, and the mean CND of the diverged sequence is 13.82 (black dot) indicating that the diverged sequences are much more likely to represent additional duplicate copies that are unrepresented reference genome (GRCh38) (two-sided Mann-Whitney test; p=2.03*10–5). The boxes indicate the range between the first and third quartiles, with the bold line specifying the median. The whiskers show the minimum and maximum within 1.5 times the interquartile range extending from the first and third quartiles. Copy number was estimated in CHM1 examining k-mer frequency found in Illumina WGS reads; methods are described in Sudmant et al. 2015. A similar approach was used for estimating copy number in the reference except we generated simulated reads using the reference and then estimated copy number in the same fashion using the simulated reads.