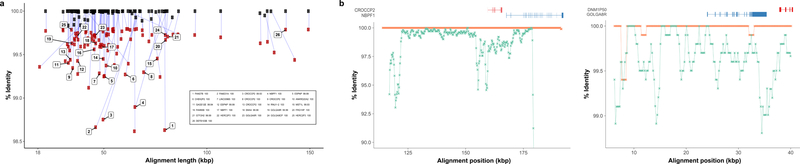
Figure 4. Correspondence between SDA sequence-diverged contigs and BACs.
a) The figure depicts the alignment length and percent identity sequence match for n=105 diverged SDA contigs compared to BAC clones (black) sequenced from the same source individual (CHM1) and the human reference genome (GRCh38) (red). The 15 most diverged sequences with respect to the reference and those containing duplicons as described by Jiang et al., 2007 and a more recent analysis of the human genome (n = 26) are shown. (See Tables S5 and S6 for more details.) b) Two examples of genes corresponding to diverged duplications are shown where the SDA sequence is aligned to both the reference genome (blue) and the CHM1 BACs (orange). BLASR alignments are computed in 1000 bp windows sliding 500 bp (steps).