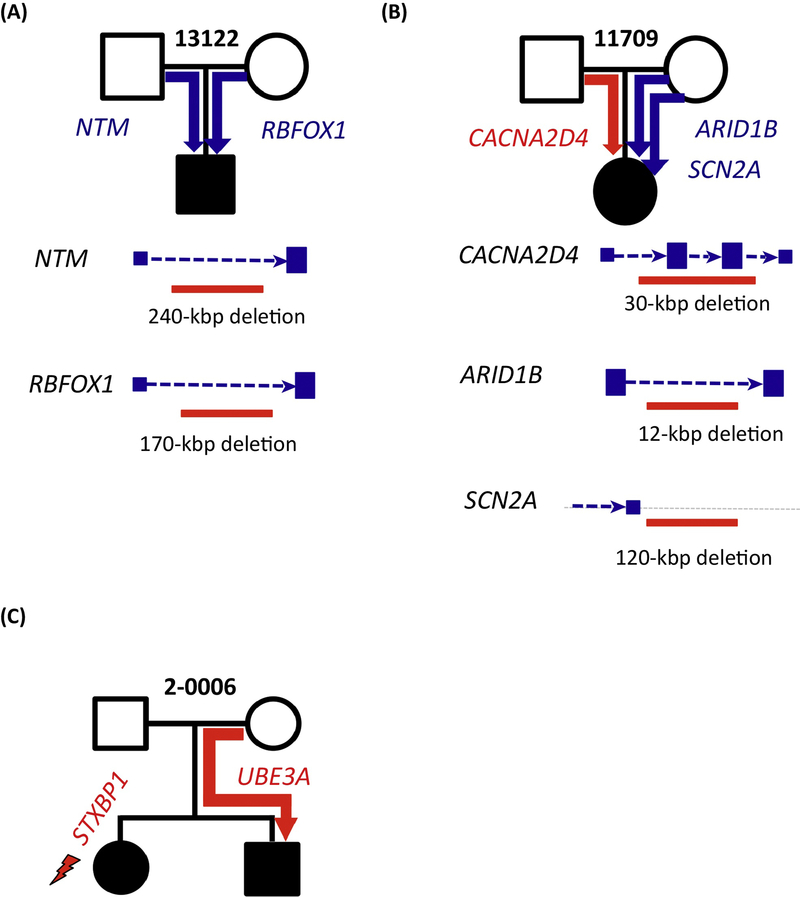
Figure 1: Genetic heterogeneity and multiple disruptive mutations in autism pedigrees.
In ~10% of patients with autism, genome sequencing reveals multiple de novo and disruptive mutations in different genes and their regulatory DNA. For example, (A) a child from SSC family 13122 inherits two large deletions affecting putative regulatory regions of autism risk genes NTM and RBFOX1. (B) Similarly, a child in SSC family 11709 inherits three different deletions with two affecting putative regulatory regions of the autism risk genes ARID1B and SCN2A. (C) Examination of families with multiple affected individuals frequently finds that a genetic risk variant segregates to only one of the two children or that different affected individuals each carry a different risk variant. For example, in MSSNG multiplex family 2–0006 [34], one autistic offspring carries a de novo loss-of- function event in STXBP1 while the other affected male sibling carries a maternally inherited loss-of-function event in UBE3A. Panels (A) and (B) are adapted from Turner et al. 2016 [17]. MSSNG family 2–0006 was studied in Yuen et al. 2015 [34].