Figure 4:
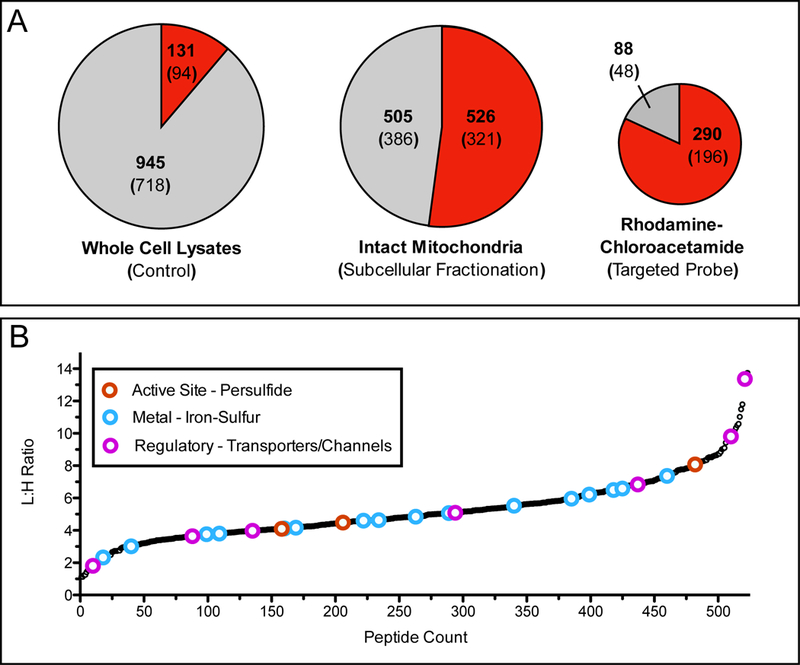
Application of organelle-specific proteomics to examine mitochondrial reactive cysteine residues. (A) Pie charts demonstrate the fraction of mitochondrial peptides identified through probe labeling and proteomic analysis of whole cell lysates by iodoacetamide (IA)-alkyne (left), enriched mitochondria labeled with IA-alkyne (center) [65], and live cells labeled with rhodamine-chloroacetamide (right) [66]. Red shading indicates fraction of total peptides annotated as mitochondrial, while the gray shading indicates peptides from other subcellular organelle or those without annotation. Bold number indicates identified peptides, number within parentheses is the total number of proteins those peptides were identified from. (B) L/H ratios for mitochondrial cysteine-containing peptides identified by labeling of enriched mitochondria with either 100 μM (light) or 10 μM (heavy) IA-alkyne. Lower L/H ratios indicate more reactive cysteine residues. Colored open circles indicate cysteine residues that are annotated with functions described previously as important and somewhat specific to the mitochondria (red - active site cysteine persulfide, blue - iron-sulfur ligating, and pink - regulatory oxidation). Importantly these annotations are generally not observed during whole cell lysate analysis.
