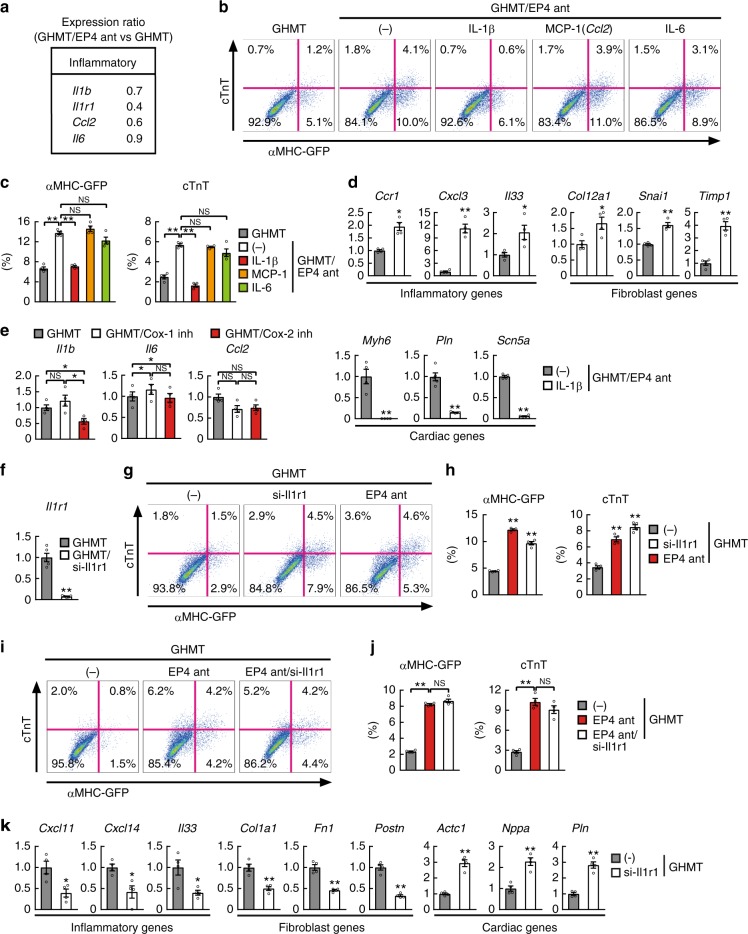
Fig. 7.
EP4 antagonist induces cardiac reprogramming via inhibition of IL-1β/IL-1R1 signaling. a The relative mRNA expression in GHMT/EP4 antagonist-TTFs compared to GHMT-TTFs by microarray. See also Fig. 6. b, c FACS analyses for αMHC-GFP+ and cTnT+ cells. GHMT-transduced TTFs were cultured with EP4 antagonist with or without IL-1β, IL-6, or MCP-1 for 1 week. Quantitative data are shown in c; n = 4 biologically independent experiments. d Relative mRNA expression in GHMT/EP4 antagonist-TTFs with or without IL-1β; n = 4 biologically independent experiments. e qRT-PCR analyses of inflammatory gene expression in postnatal TTFs treated with COX inhibitors for 1 week; n = 4 biologically independent experiments. f Relative mRNA expression of Il1r1 in GHMT-TTFs transfected with scrambled siRNA or si-Il1r1; n = 5 biologically independent experiments. g, h FACS analyses for αMHC-GFP+ and cTnT+ cells. GHMT-transduced TTFs were cultured with or without si-Il1r1 or EP4 antagonist (ant) for 1 week. Quantitative data are shown in h; n = 4 biologically independent experiments. i, j FACS analyses for αMHC-GFP+ and cTnT+ cells. GHMT-transduced TTFs were cultured with EP4 antagonist (ant) with or without si-Il1r1 for 1 week. Quantitative data are shown in j; n = 4 biologically independent experiments. k Relative mRNA expression of inflammatory genes, fibroblast genes, and cardiac genes in GHMT-TTFs transfected with scrambled siRNA or si-Il1r1; n = 4 biologically independent experiments. Student’s t-test was performed for d, f, k, one-way ANOVA with Dunnett’s post hoc test was performed for h, one-way ANOVA with Tukey’s post hoc test was performed for c, e, j; all data are presented as mean ±SEM. *P < 0.05, **P < 0.01 vs. the relevant control. NS, not significant