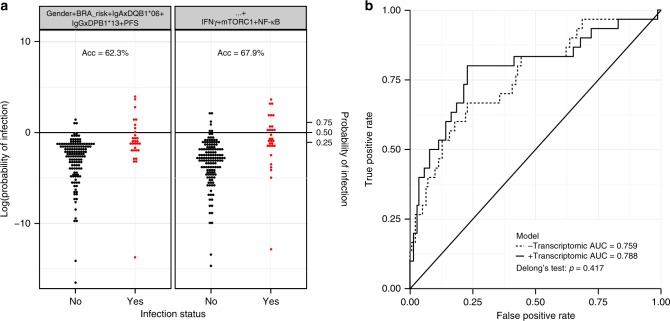
Fig. 4.
Prediction of the response does not improve by adding transcriptomic data. a Logistic regression models were built to predict HIV infection status of RV144 vaccinees (142 vaccinees that were HIV negative at last follow-up and 30 vaccinees that acquired HIV). The accuracy of each model was assessed by tenfold cross-validation. The first model (left panel) included IgA against V2, IgG against V1/V2, and the polyfunctionality score (PFS) previously identified as markers of response to RV144 vaccine. The second model (right panel) included the same markers as the first model but with the addition of the three pathways associated with HIV status in the transcriptomic analysis (IFNγ response, MTORC1 signaling, and TNFα signaling via NF-κB). The balanced accuracy (Acc) of each model is given on the plot. b Corresponding ROC curves based 10-fold cross-validation for the model without the three genesets and the model with the three genesets identified in the transcriptomic analysis