Figure 5.
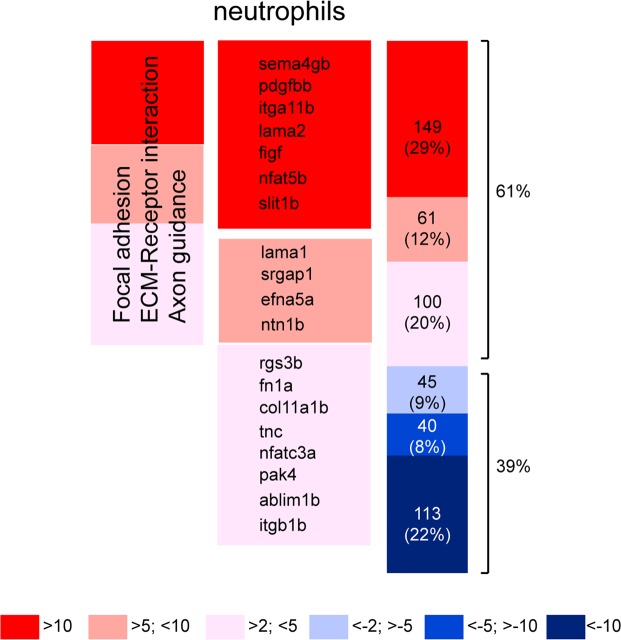
Cxcr4b transcriptomic signature in zebrafish neutrophils. (A) Heatmap showing up- and down-regulated genes in cxcr4b−/− neutrophils compared to cxcr4b+/+ neutrophils. 61% is the percentage of up-regulated genes, whereas 39% is the percentage of down-regulated genes. Genes involved in focal adhesion, ECM-Receptor interaction and axon guidance are up-regulated. Percentages of up- or down-regulation are calculated based on the total number of genes left after a cutoff of p < 0.05 in both DESeq and edgeR (neutrophil dataset: from n = 21517 to n = 508). Before the analysis, 107 genes were manually removed in the neutrophil dataset due to high variation among the triplicates. An alternative analysis method using DESeq2 paired confirmed the affected pathways (Table 2).