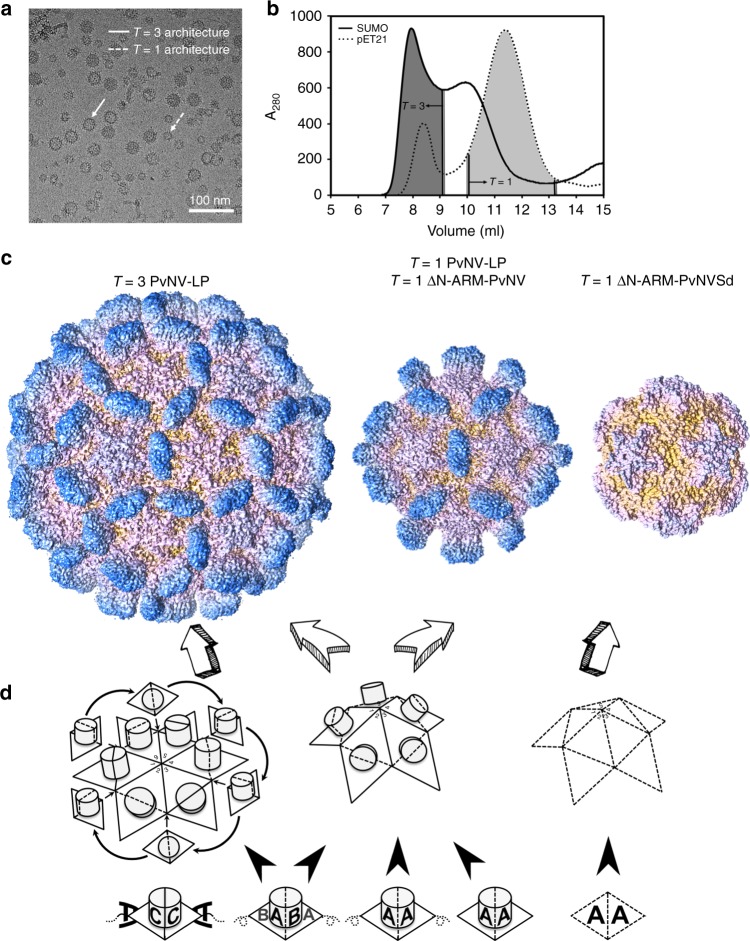
Fig. 6.
Particle polymorphism and self-assembly mechanisms of shrimp nodavirus. a Cryo-EM image of the PvNV-LP. The diameters of the PvNV-LPs are near 35 and 24 nm, corresponding to the T = 3 and T = 1 particles. T = 3 and T = 1 capsids are indicated by white solid and dotted arrows, respectively. Scale bar = 100 nm. b The elution profiles of the PvNV-LP and pET21-PvNV-LP in the size-exclusion chromatography. The various fractions were collected and their absorbance at 280 nm were measured. c Cryo-EM, X-ray crystal and EM structures of the T = 3 and T = 1 PvNV-LPs, T = 1 ΔN-ARM-PvNV SVP and T = 1 ΔN-ARM-PvNVSd SVP. The T = 3 PvNV-LP surface and two T = 1 SVPs surfaces are rainbow-colored according to their distance from the spike to shell. d The diagram shows the putative self-assembly process of the T = 3 and T = 1 shrimp nodavirus capsids. The basic unit of capsid assembly is the dimeric capsomeres (diamond with black solid and dotted lines). The N-ARM deletion and the N-terminal fusion tag both might guide the assembly of the T = 1 capsid