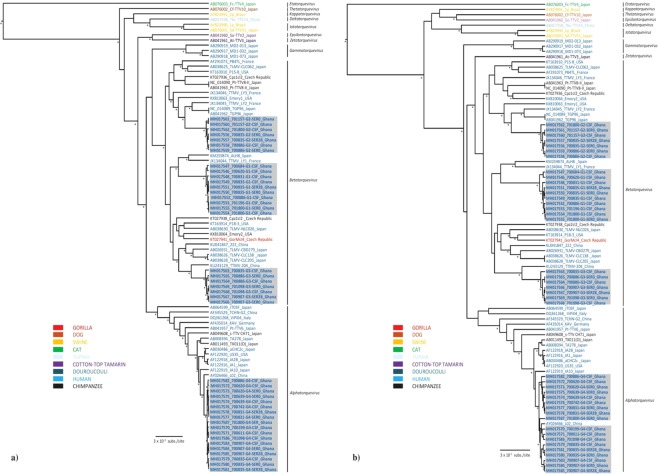
Figure 5.
Bayesian maximum clade credibility (MCC) trees based on complete nucleotide (a) and amino acid (b) sequences of ORF1 showing the phylogenetic placement of the TTMV-G1-G3 and TTVG4 species from this study compared with representative members of the Betatorquevirus, Alphatorquevirus and other genera of the Anelloviridae family. Bayesian posterior probabilities (≥90%) and parallel maximum likelihood replicate percentages (≥70%) in which the associated taxa clustered in the bootstrap test (1,000 replicates) are indicated at the nodes (asterisk). The main lineages/genera are indicated to the right of the tree. Boldface indicates TTMV and TTV species from febrile pediatric patients (this study). Taxon information includes strain names, GenBank accession numbers, and countries of origin. The TTV strains are colored according to the host species. Scale bar indicates mean number of nucleotide/amino acid substitutions per site.