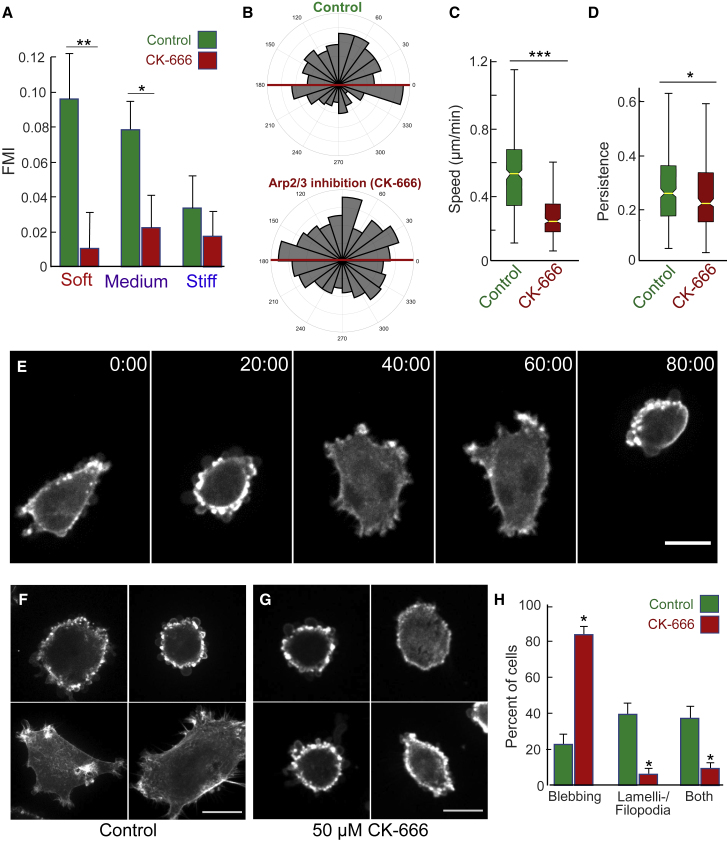
Figure 8.
Impaired durotaxis resulting from inhibition of Arp2/3. (A) FMI for cells treated with 50 μM CK-666 (red) or DMSO vehicle control (green) moving parallel to the stiffness gradient within the soft, medium, or stiff regions. (B) Angular displacement analysis reveals no evidence for a substrate-directed response of CK-666 treated cells (bottom) cultured on a stiffness gradient compared to vehicle control cells (top). (C) Speed and (D) persistence (directionality) of cell migration are reduced with CK-666 treatment. Data for 320 DMSO- and 391 CK-666-treated cells from N ≥ 3 biological replicates (independent experiments) were collected. (E) Time series of an individual U87 glioblastoma cell (untreated) that spontaneously alternates phenotype periodically between lamellipodial-filopodial and blebbing. (F) Representative fluorescence images of control (untreated) U87 glioblastoma cells expressing TagGFP2-LifeAct to label F-actin showing phenotypes varying from rounded and blebbing to poorly polar cells with multiple small lamellipodia and short spike-like filopodia. (G) Representative fluorescence images of U87 glioblastoma cells treated with 50 μM CK-666, often showing numerous small blebs at the cell periphery. (H) Morphological quantification of control (green bars) versus CK-666-treated (red) U87 cells with blebbing, lamellipodial-filopodial, or both phenotypes in time-lapse videos. (H) FMI p-values were calculated using one-way ANOVA to compare between groups. (∗∗∗p ≤ 0.001, ∗∗p ≤ 0.01, ∗p ≤ 0.05). Angular displacement data were analyzed statistically using Rayleigh tests for evaluating circular data with the assumed mean direction set at 90° (∗∗∗p ≤ 0.001). In (A) and (H), error bars represent mean ± SE. In (C) and (D), boxplot center lines denote the median and edges represent the 25th and 75th percentiles, with whiskers incorporating 99.3% of all data. Scale bars, 10 μm for (E) and 20 μm for (F) and (H). To see this figure in color, go online.