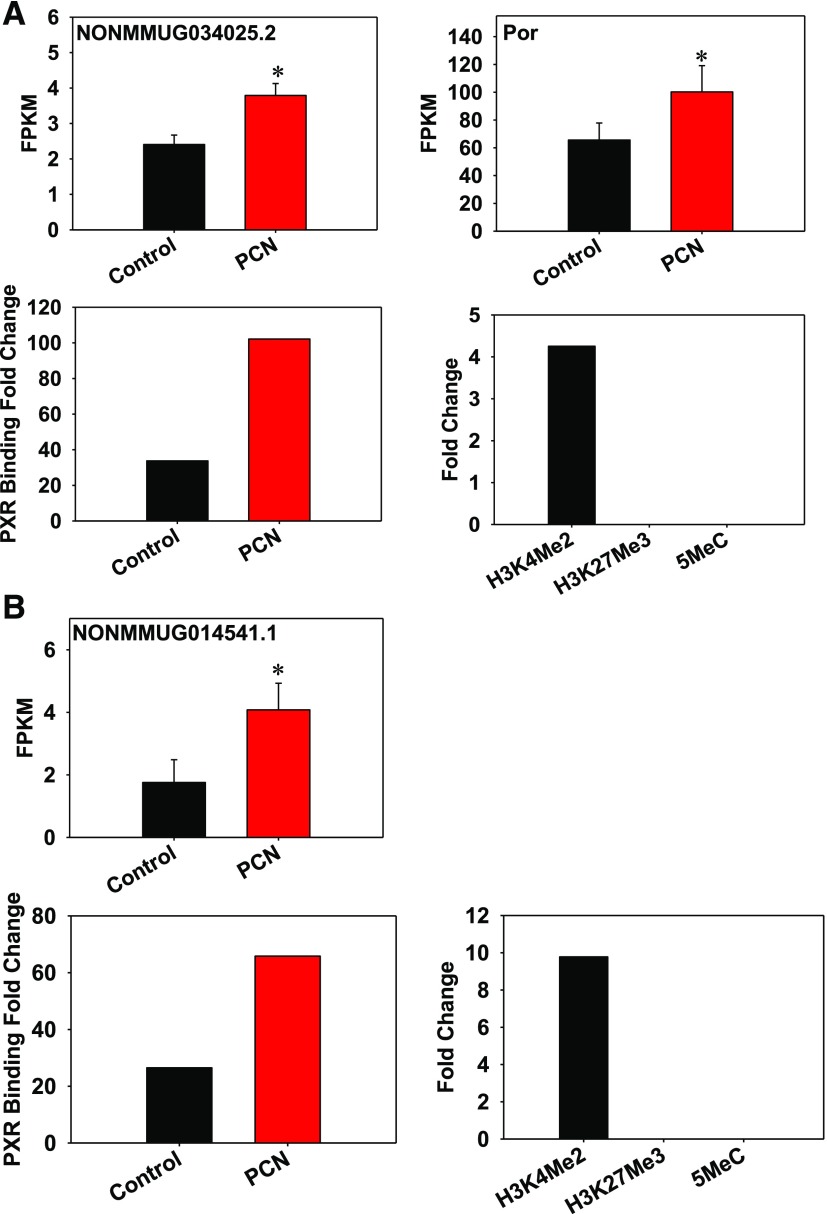
Fig. 5.
(A) Colocalization of PXR and H3K4me2 around the lncRNA NONMMUG034025.2 and the paired PCG Por gene loci. Integrated Genome Viewer (IGV) is a high-performance visualization tool for displaying and exploring large data sets, including RNA-Seq data, and can be accessed at http://software.broadinstitute.org/software/igv/. The genomic locations of lncRNA and PCG are visualized by IGV. Asterisks represent statistically significant differences compared with corn oil control group (P < 0.05, Cuffdiff). FPKM is a unit to express RNA abundance from the RNA-Seq data. Data from RNA-Seq (corn oil– and PCN-exposed groups), ChIP-Seq (for PXR-DNA binding in corn oil– and PCN-exposed conditions), and ChIP-on-chip (for H3K4me2, H3K27me3, and 5MeC on mouse chormosomes 5, 12, and 15) were integrated as described in Materials and Methods. (B) Colocalization of PXR and H3K4me2 around the intergenic lncRNA NONMMUG014541.1 gene locus. The genomic location of lncRNA and PCG is seen by IGV. Asterisks represent statistically significant differences compared with corn oil control group (P < 0.05, Cuffdiff). Data from RNA-Seq (corn oil– and PCN-exposed groups), ChIP-Seq (for PXR-DNA binding in corn oil– and PCN-exposed conditions), and ChIP-on-chip (for H3K4me2, H3K27me3, and 5MeC on mouse chormosomes 5, 12, and 15) were integrated as described in Materials and Methods.