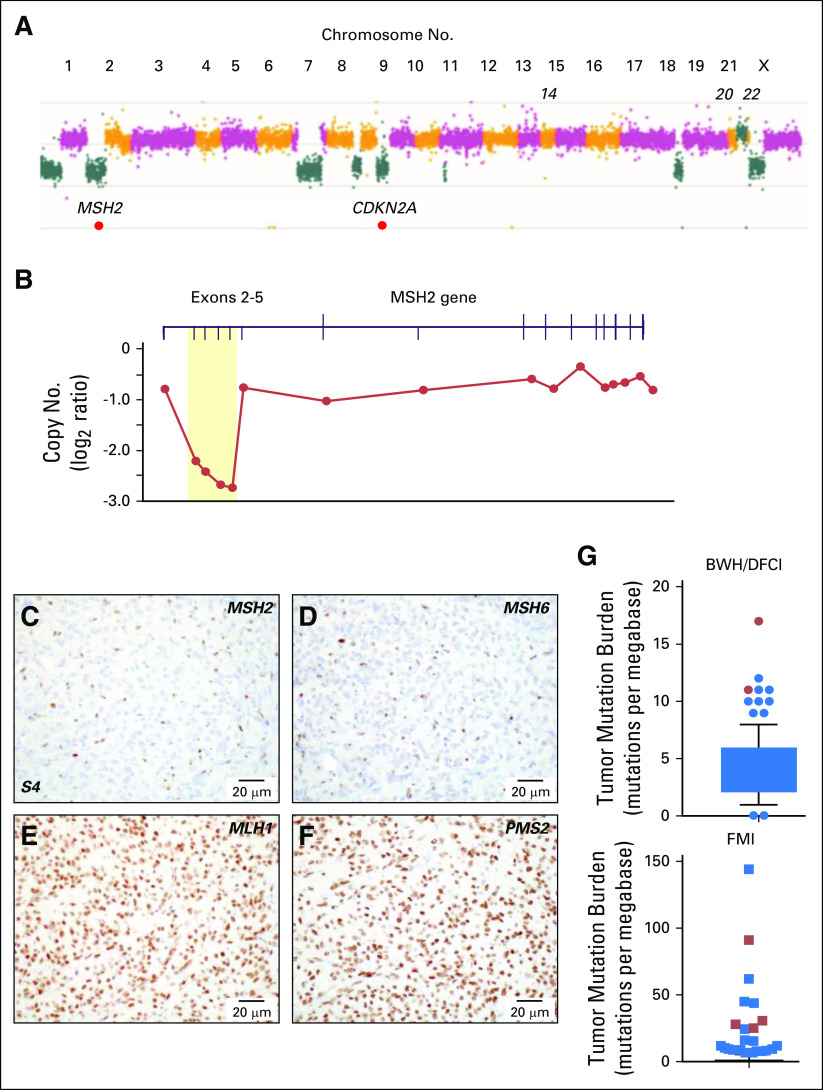
Fig 3.
Genomic and immunohistochemical characterization of meningioma samples. (A) Copy number analysis from targeted next-generation sequencing data from sample 6 identified a genome-wide profile characteristic of a high-grade meningioma, including loss of 1p, 9p and monosomy of chromosomes 18 and 22. Focal homozygous deletion of CDKN2A/CDKN2B and intragenic deletion of MSH2 were present. (B) Gene-level view of MSH2 showed a homozygous intragenic deletion of exons 2 through 5 of MSH2 (NM_000251; log2 ratio from −2.21 to −2.73) in the setting of 2p arm-level single copy loss. Copy number is depicted as a log2 ratio value. Immunohistochemistry on pretreatment meningioma resection (surgery 4/sample 4; S4) for (C) MSH2, (D) MSH6, (E) MLH1, and (F) PMS2. MSH2 and MSH6 staining was present only in nontumor cells. (G) Box and whiskers plot (5th to 95th percentiles) of tumor mutational burden (TMB; mutations per megabase) for Brigham and Women’s Hospital (BWH)/Dana-Farber Cancer Institute (DFCI) profile initiative cohort (n = 228 sequenced cases; Data Supplement) plus sequencing data for the mismatch repair (MMR)–deficient case BWH/DFCI 2 and for the Foundation Medicine cohort (FMI; n = 615 sequenced cases; Data Supplement). Patient cases with loss of function changes in MMR and MMR-related genes (from Table 2) are highlighted in red (dots and squares). For the BWH/DFCI profile initiative cohort, the mean TMB was 4.25 mutations/megabase (standard deviation, 2.55; standard error of the mean, 0.17; lower 95% CI of mean, 3.91; upper 95% CI of mean, 4.58). For the Foundation Medicine cohort, the mean TMB was 2.77 mutations/megabase (standard deviation, 8.08; standard error of the mean, 0.33; lower 95% CI of mean, 2.14; upper 95% CI of mean, 3.41).