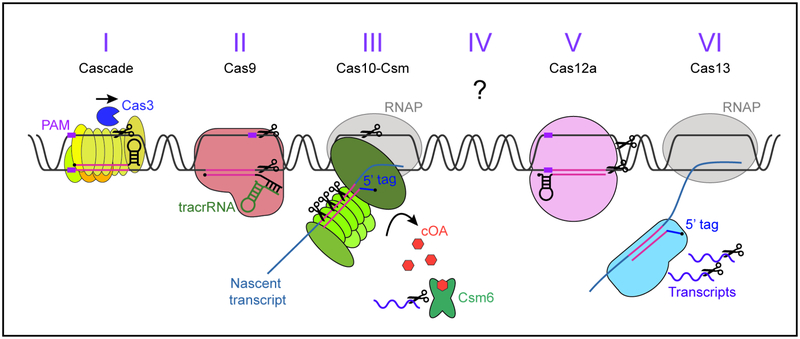
Figure 3. Different CRISPR targeting mechanisms.
See text for details. Purple box: PAM, black circles: crRNAs 5’ end, pink: spacer/protospacer sequences, blue: 5’ crRNA tags inhibiting type III/VI autoimmunity. For types I, II, and V, the DNA double helix is unwound by the main effector complex in a PAM-dependent manner, and DNA is cut by Cas3 (type I) or Cas9/Cas12a (types II/V). Type III and VI recognise the protospacer within a nascent transcript in a PAM-independent manner, this is followed by the cleavage of DNA and/or RNA from the invader. Type III also produces cyclic oligoadenylates (orange hexagons) which allosterically activate the accessory RNase Csm6 (Csx1 for III-B).