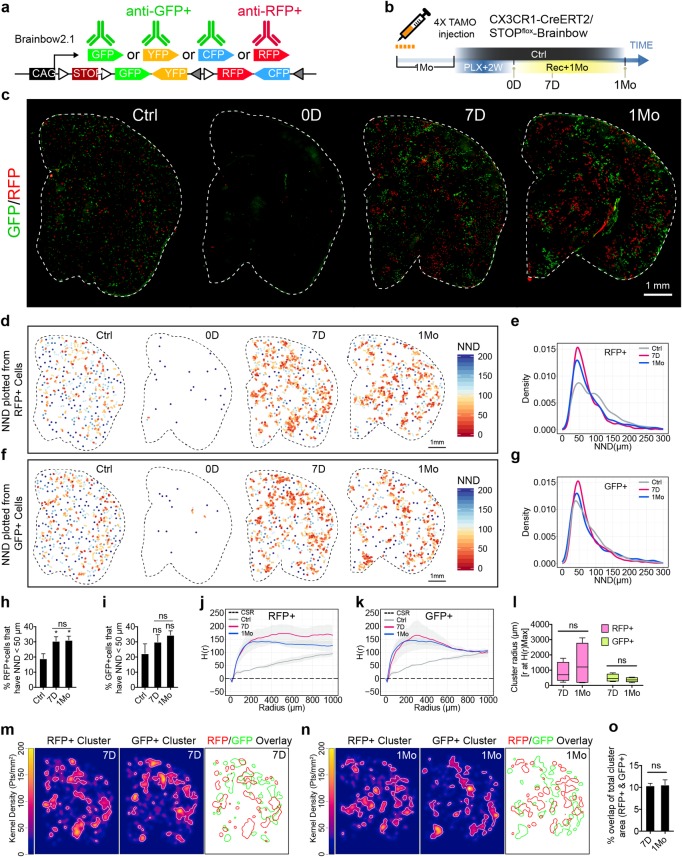
Fig 4. Repopulated microglia form stable clusters with minimal migratory diffusion.
(a) Immunofluorescent staining strategy to visualize RFP+ or GFP+ microglia using the Brainbow reporter. (b) Experimental scheme to sparsely label microglia with Brainbow reporter. The CX3CR1-CreERT2/STOPflox-Brainbow mice (7–8 Mo) were given a daily dose of 2 mg tamoxifen per animal via IP injection for 4 days. Labeled mice were subjected to 2 weeks of PLX treatment before switching to normal diet for 7 D or 1 Mo. (c) Representative images from coronal sections of RFP+ cells (red) and GFP+ cells (green). Parenchyma outline is visualized with white dotted line. (d) Spatial heatmap of NND reconstructed from RFP+ cells. Each dot represents a single cell color-coded by NND score. (e) Density plot showing NND distribution from RFP+ cells. (f) Spatial heatmap of NND reconstructed from GFP+ cells. (g) Density plot showing NND distribution from GFP+ cells. (h) Quantification of the percentage of RFP+ cells that have equal or less than 50 μm NND (mean ± SEM). Animals used: Ctrl (n = 5), 7 D (n = 4), and 1 Mo (n = 5). One-way ANOVA with Dunnett's multiple comparisons test was used by comparing to the Ctrl group. Sidak's multiple comparisons test was used to compare 7 D and 1 Mo. (i) Quantification of the percentage of GFP+ cells that have equal or less than 50 μm NND. Statistical analysis was the same as (h). (j, k) Plot of Ripley’s H-function analysis on RFP+ (h) and GFP+ (i) cell-clustering patterns. Black dotted line represents CSR, i.e., absence of clustering pattern. Average H(r) value from each group were plotted. Grey ribbon shades represent SEM. Animals used: Ctrl (n = 5), 7 D (n = 4), and 1 Mo (n = 5). (l) Cluster domain size estimation from H(r)Max. Box-whisker plot of cluster domain size estimation from H(r)Max (whisker: max and min; box: 25 and 75 percentile). Unpaired t test was used. (m, n) 2D kernel density map showing cluster interaction between RFP+ and GFP+ cells. Representative sample from 7 D (m) and 1 Mo (n) were plotted. White line marks the border of isolated cluster domains based on the top 10% of the highest kernel density. Overlay of the isolated RFP+ and GFP+ cluster contours are delineated with red and green lines, respectively. (o) Quantification of the percentage of overlapping area of RFP+ and GFP+ clusters with respect to total RFP+ and GFP+ cluster area (mean ± SEM). Animals used: 7 D (n = 4); 1 Mo (n = 5). Unpaired t test was used. P value is summarized as ns (P > 0.05), *(P ≤ 0.05), **(P ≤ 0.01), ***(P ≤ 0.001), and ****(P ≤ 0.0001). Individual numerical values can be found in S1 Data. CreERT2, tamoxifen-inducible Cre recombinase; CSR, complete spatial randomness; Ctrl, control; CX3CR1, CX3C chemokine receptor 1; D, days; GFP, green fluorescent protein; IP, intraperitoneal; Mo, months; NND, nearest neighbor distance; PLX, PLX5622; RFP, red fluorescent protein.