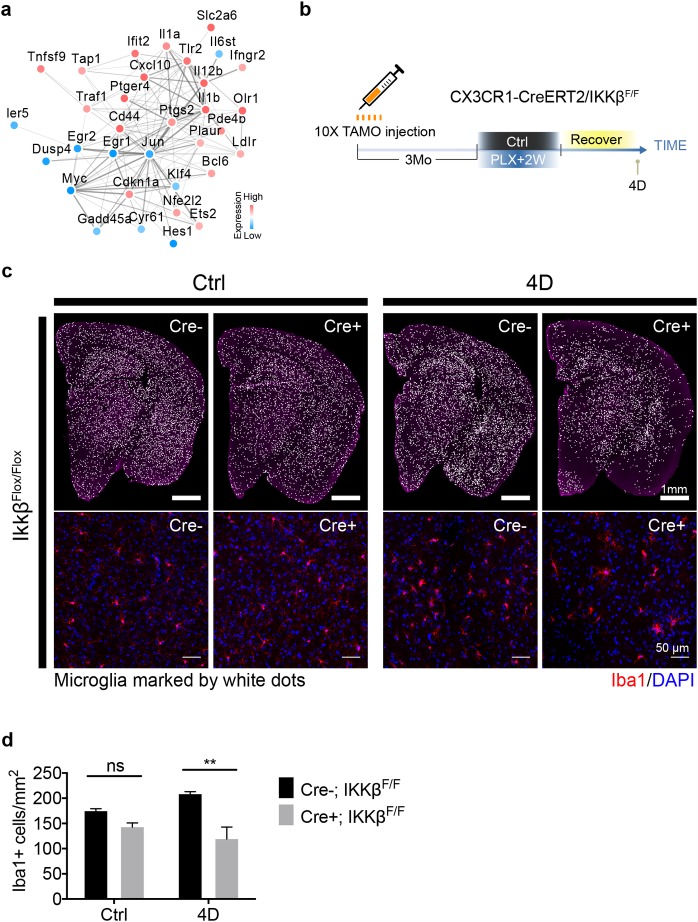
Fig 7. NF-kβ pathway promotes microglia repopulation.
(a) Network of NF-kβ signaling–associated genes identified from the RNA-seq experiment (fast-return genes). Network was constructed using the STRING database. Directionality of gene expression pattern is shown as red for up-regulation and blue for down-regulation. (b) Experimental scheme for generating conditional IKKβ knockout mice. CX3CR1-CreERT2/IKKβf/f mice (7–8 Mo) injected with 2 mg tamoxifen/day via IP for 10 days. Three months later, mice were treated with PLX5622 diet for 2 weeks and switched to a normal diet for 4 D. (c) Representative images showing microglial density from the whole coronal section (upper panel) and thalamic region (lower panel). Iba1+ cells are visualized as white dots. (d) Quantification of microglial density from entire coronal section as shown in panel (c, upper panel) (mean ± SEM). Animal used: (n = 3) for each group. Two-way ANOVA with Sidak's multiple comparisons test was used to compare IKKβ control (Cre−) and IKKβ deletion (Cre+). P value is summarized as ns (P > 0.05), *(P ≤ 0.05), **(P ≤ 0.01), ***(P ≤ 0.001), and ****(P ≤ 0.0001). Individual numerical values can be found in S1 Data. CreERT2, tamoxifen-inducible Cre recombinase; CX3CR1, CX3C chemokine receptor 1; D, days; IKKβ, I-Kappa-B-Kinase Beta; IP, intraperitoneal; Mo, months; PLX, PLX5622; STRING, Search Tool for the Retrieval of Interacting Genes/Proteins.