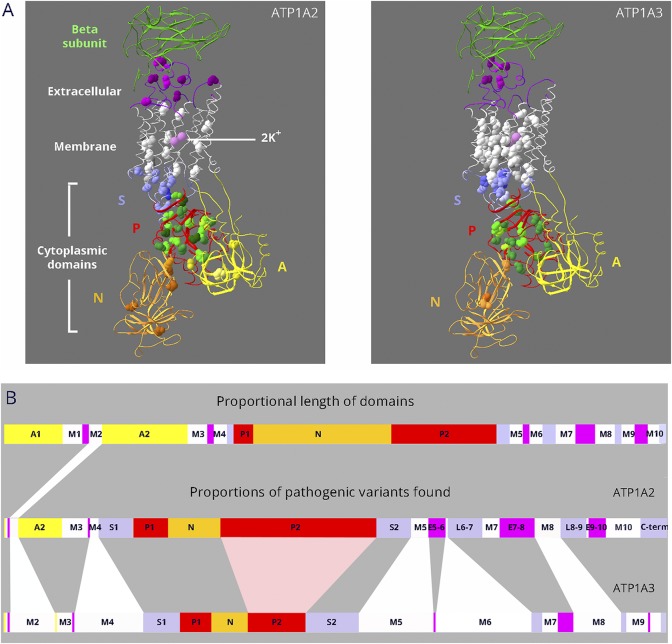
Figure 1. Dissimilar mutation distributions in 88% identical ATP1A2 and ATP1A3 proteins.
(A) Ribbon diagrams of Na,K-ATPase in the K+-bound conformation. Bound K+ is shown as pink spheres. Extracellular portions include the β-subunit (green; transmembrane portion removed for clarity) and extracellular loops of the α-subunit (magenta). The 10 transmembrane α-helices are white. The cytoplasmic domains are the stalk (S, lavender), phosphorylation domain (P, red), nucleotide binding domain (N, gold), and actuator domain (A, yellow). Spacefill residues are the backbones of each amino acid mutated in ATP1A2 or ATP1A3. Color shading is varied to help distinguish different amino acids. For ATP1A2, 76 different DNA variants occur in 66 amino acids, i.e., 10 amino acids have 2 alternative codon changes. Three of the mutations are single amino acid deletions. For ATP1A3, 130 different DNA variants occur in 86 amino acids; 5 are single amino acid deletions, and there are 25 amino acids with 2–6 alternative codon changes. All 18 overlapping residue pairs (including 3 where one was found in ClinVar) were identical before the mutation. In 6 pairs, the amino acid change was different; in 8, it was the same; and in 4, both identical and different substitutions were found (i.e., alternative codon changes). (B) Linear diagram of the domain substructure. Both the A domain (yellow) and the P domain (red) are composed of 2 parts that are separated in the linear structure. As in (A), the extracellular loops are magenta. The stalk domain comprises 5 separate parts: the cytoplasmic extensions of the M4 and M5 transmembrane spans S1 and S2; the short intracellular loops L6-7 and L8-9; and the C-terminus (lavender). For ATP1A2 and ATP1A3, domain lengths are proportional to the number of distinct variants found (including alternative codon changes). Although variants are found in most domains, ATP1A2 has an excess in the P domain, and ATP1A3 has an excess in the membrane domain.