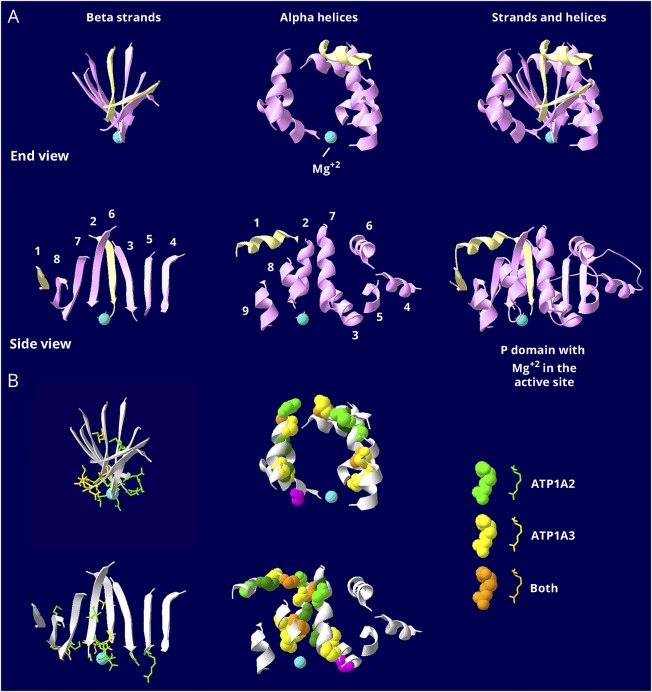
Figure 3. Similar but restricted distributions of pathogenic variants in the P domain.
The P domain is the most conserved in the gene family. It is covalently phosphorylated during transport, and the Mg2+ ion (aqua) is at the active site. (A) The P domain consists of 2 halves that are separated in the linear structure but that interdigitate in a complex way when the protein folds. The first half, P1 in figure 1B, is shown in pale yellow, and the second half, P2 in figure 1B, in pink. There is a twisted β-sheet (left views) surrounded by α-helices (middle views). Strands and helices are numbered in the order found in the sequence. (B) The known mutations are displayed with color coding as indicated. Stick view is used for mutations in the β-sheet to avoid overcrowding. Note how most of the side chains of the mutated residues extend down into the active site surface. Spacefill view of the mutations, without side chains, is used for the α -helices. Note how the mutations are confined to just 5 helices: helix 1, which is associated with the stalk domain, and 4 helices that form a belt around the middle of the P domain: 2, 3, 7, and 8. There are no mutations in the end strands (1 and 4) or the end helices (4, 5, 6, and 9), which may be less important to function. The magenta mutation in an α-helix is the only helix mutation near (but not in) the active site. It is ATP1A3 T613M, a recurrent RDP mutation. The data indicate that the ATP1A2 and ATP1A3 distributions of mutations are very similar in the P domain, but there are 2 types of mutations. Most of the mutations in the β-sheet are at the active site. Most of the ones in the α-helices are distant from the active site. There was no correlation with phenotype severity.