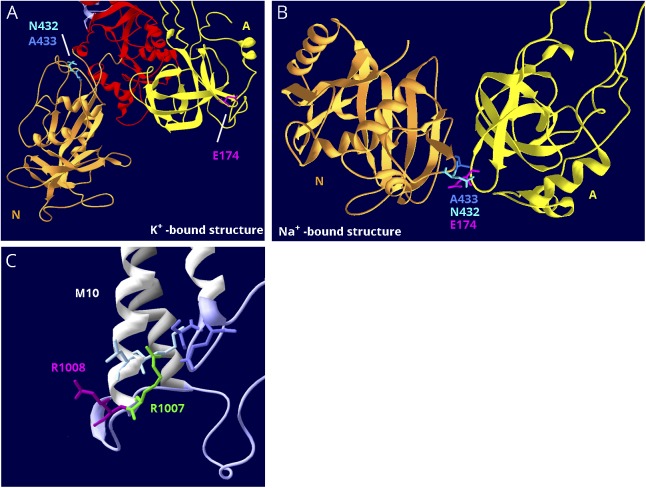
Figure 6. Structure-based interpretation of variants of uncertain significance.
(A) E174K; N, P, and A domains are visible. In the K+-bound conformation, the N (gold) and A (yellow) domains are far apart. (B) In the Na+-bound conformation, the N and A domains are in contact. The P domain in the background was hidden for clarity. E174K (magenta) is a variant in ATP1A2 that has the poorest conservation score in figure 5B, and it is predicted to be tolerated by SIFT.38 In the K+-bound structure, it is fully exposed to the cytoplasm (A), but in the Na+-bound structure, it is the point of closest contact between the A domain (yellow) and N domain (gold). There it interacts with 2 adjacent residues, an asparagine N432 and an alanine A433 (blue and aqua). They are also shown in (A). (C) R1008W; close-up of M10, M7, and M8 where these helices terminate in parts of the S domain (lavender). R1008W (magenta) is a variant in ATP1A2 in a patient with compound heterozygosity with R548C. Because the adjacent variant R1007W (green) is known to be pathogenic, it was suggested that R1008W might contribute to the severity.39 R1007 has a better conservation score, but R1008's score is still in the range of other pathogenic variants (close to zero). In the structure, R1007 points into the protein where it makes close association with 2 domains: two residues in M10 (light blue) and 2 residues in L7-8 (lavender), including the protein kinase A phosphorylation site, S933. In contrast, R1008 points out into the cytoplasm, where a substitution may be tolerated.