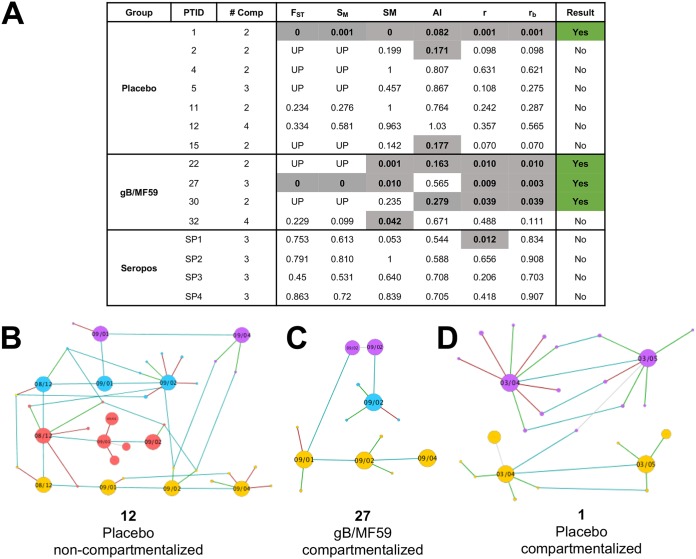
FIG 4.
Evidence of viral genetic compartmentalization at gB locus in 3 of 4 gB vaccinees. (A) Table indicating the results of 6 distinct tests of genetic compartmentalization performed on the pool of unique gB haplotypes identified per patient, including Wright’s measure of population subdivision (FST), the nearest-neighbor statistic (Snn), the Slatkin-Maddison test (SM), the Simmonds association index (AI), and correlation coefficients based on distance between sequences (r) or number of phylogenetic tree branches (rb). For each test, >1,000 permutations were simulated. Significant test results suggesting genetic compartmentalization are shown in gray with bold text. Values for FST, Snn, SM, r, and rb represent uncorrected P values, with P < 0.05 considered significant. An AI of <0.3 was considered a significant result. The presence of three or more positive tests per patient was considered strong evidence for genetic compartmentalization, indicated in green. UP, underpowered (fewer than 5 haplotypes were present in each compartment, making FST and Snn error prone). (B to D) Networks of unique viral haplotypes by individual patient, with 1 patient lacking tissue compartmentalization (B) and 2 patients demonstrating strong evidence of viral genetic compartmentalization (D). Samples are organized chronologically from left to right, with blood shown in red, saliva in blue, urine in yellow, and vaginal fluid in purple. The size of each node reflects the relative prevalence of each haplotype. Light blue lines connect identical viral variants between time points and compartments, green lines connect variants with a synonymous mutation, and red lines those with a nonsynonymous mutation.