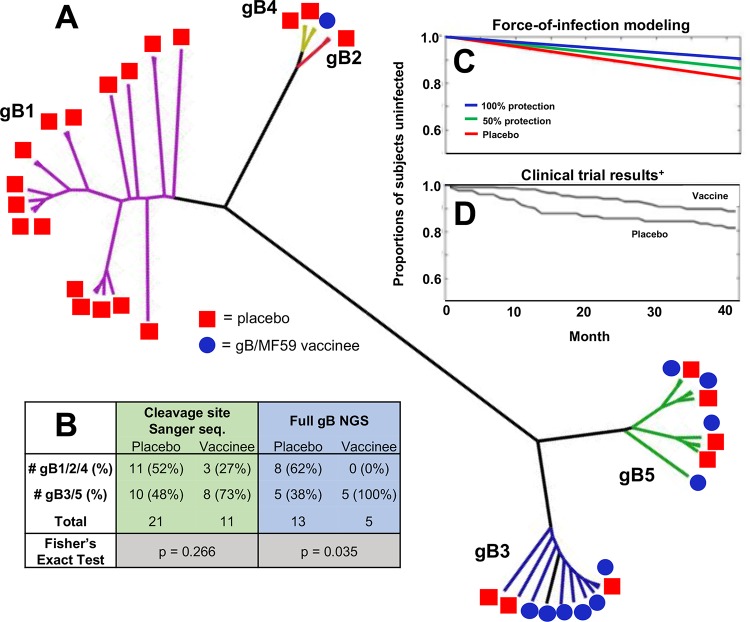
FIG 5.
Possible protection against gB1/2/4 genotype supergroup viruses in gB/MF59 vaccinees. (A) Unrooted phylogenetic tree in a polar layout constructed using full gB open reading frame consensus sequences for each sequenced sample. Note, a consensus sequence was not defined for all samples from each subject, so this tree depicts multiple samples representing multiple time points/compartments for each trial participant. Samples with discrepant genotypes from Sanger sequencing not excluded. Clades representing gB genotypes are highlighted in different colors: gB1, purple; gB2, yellow; gB3, blue; gB4, red; gB5, green. (B) Numbers of distinct gB vaccinees and placebo recipients who acquired viral variants belonging to two supergroups of genetically similar gB genotypes (gB1/2/4 and gB 3/5) as defined by Sanger sequencing of the cleavage site (highlighted in green) and NGS of the full gB protein (highlighted in blue). (C) Force-of-infection modeling closely predicts observed gB/MF59 vaccine trial efficacy (D). Force-of-infection model iterations indicate that gB vaccinees are universally protected against acquisition of gB1/2/4 variants (blue line; consistent with full gB NGS data) or 50% protected (green line; consistent with Sanger sequencing data). Model makes assumption that (i) viruses belonging to the gB1/2/4 genotype supergroup represent 52% of the circulating virus pool and (ii) the HCMV force of infection is 5.7 per 100 person-years. (Panel D adapted from The New England Journal of Medicine [5].)