FIG 2.
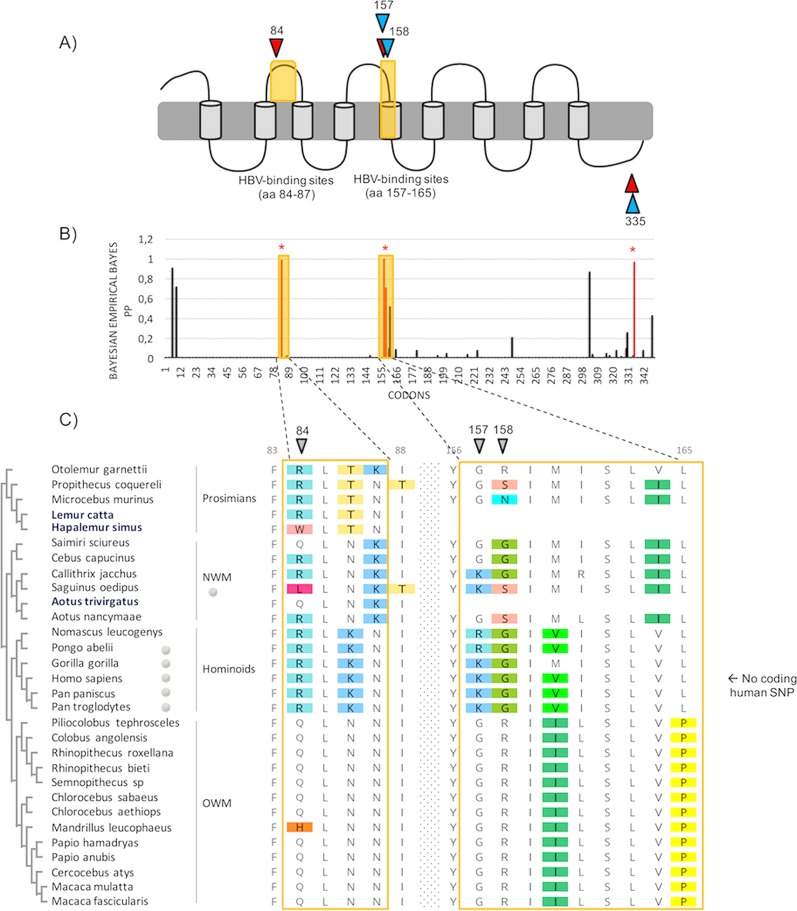
Primate NTCP has evolved under positive selection, with positively selected sites mapping to the HBV-binding region. (A) Representation of NTCP with positively selected sites. A diagram representing the predicted domains of NTCP is shown (89), and the HBV-binding interfaces are highlighted in yellow. Specific codons found to be evolving under positive selection (PAML, Bayesian empirical Bayes [BEB] posterior probability [PP] of >0.9; MEME, P value of <0.1; FUBAR, PP of >0.9; REL, Bayes factor [BF] of >80), by at least three of the models used in this study (see Materials and Methods), are indicated in red and in blue for the primate (n = 27) and the simian primate (n = 24) data sets, respectively. aa, amino acids. (B) Histogram of the BEB PPs for a dN/dS ratio of >1 at each codon in an alignment of primate NTCP coding sequences. BEB values were inferred under the M8 site model implemented in PAML Codeml. The codons shown with stars are those identified as being under positive selection by at least three methods. (C) Amino acid alignment of the HBV-binding regions of NTCP from primates (27 publicly available species and 3 de novo-sequenced species). On the left, the cladogram of primate NTCP, with the newly sequenced species highlighted in blue, is presented. Amino acid alignment was performed with PRANK, and the color-coding is from Rasmol (Geneious, Biomatters). The positively selected sites, inferred by at least three methods, are indicated by the gray symbols. On the right, human SNP information is given for the HBV-binding regions in human NTCP. NWM and OWM, New and Old World monkeys, respectively. Codon numbering is based on the human NTCP sequence as a reference.