Uncapped viral RNAs often rely on their 5′ leader sequences to initiate translation, and the Triticum mosaic virus (TriMV) devotes an astonishing 7% of its genome to directing ribosomes to the correct AUG. Here we uncover a novel mechanism by which a TriMV cis-regulatory element controls cap-independent translation. The upstream region of the functional AUG contains a 16-nt polypyrimidine tract located 11 nt from the initiation site. Based on functional redundancy with similar motifs derived from human picornaviruses, the motif is likely to operate by directing ribosome targeting through base pairing with 18S rRNA. Our results provide the first report of a broad-spectrum mechanism regulating translation initiation for both plant- and animal-hosted picornaviruses.
KEYWORDS: 5′ UTR, IRES, YX-AUG, cap independent, plant virus, potyvirus, translation
ABSTRACT
Several viruses encode an internal ribosome entry site (IRES) at the 5′ end of their RNA, which, unlike most cellular mRNAs, initiates translation in the absence of a 5′ m7GpppG cap. Here, we report a uniquely regulated translation enhancer found in the 739-nucelotide (nt) sequence of the Triticum mosaic virus (TriMV) leader sequence that distinguishes the preferred initiation site from a plethora of IRES-encoded AUG triplets. Through deletion mutations of the TriMV 5′ untranslated region (UTR), we show that the TriMV 5′ UTR encodes a cis-acting picornaviral Y16-X11-AUG-like motif with a 16-nt polypyrimidine CU-tract (Y16), at a precise, 11-nt distance (X11) from the preferred 13th AUG. Phylogenetic analyses indicate that this motif is conserved among potyviral leader sequences with multiple AUGs. Consistent with a broadly conserved mechanism, the motif could be functionally replaced with known picornavirus YX-AUG motifs and is predicted to function as target sites for the 18S rRNA by direct base pairing. Accordingly, mutations that disrupted overall complementarity to the 18S rRNA markedly reduced TriMV IRES activity, as did the delivery of antisense oligonucleotides designed to block YX-AUG accessibility. To our knowledge, this is the first report of a plant viral IRES YX-AUG motif, and our findings suggest that a conserved mechanism regulates translation for multiple economically important plant and animal positive single-stranded RNA viruses.
IMPORTANCE Uncapped viral RNAs often rely on their 5′ leader sequences to initiate translation, and the Triticum mosaic virus (TriMV) devotes an astonishing 7% of its genome to directing ribosomes to the correct AUG. Here we uncover a novel mechanism by which a TriMV cis-regulatory element controls cap-independent translation. The upstream region of the functional AUG contains a 16-nt polypyrimidine tract located 11 nt from the initiation site. Based on functional redundancy with similar motifs derived from human picornaviruses, the motif is likely to operate by directing ribosome targeting through base pairing with 18S rRNA. Our results provide the first report of a broad-spectrum mechanism regulating translation initiation for both plant- and animal-hosted picornaviruses.
INTRODUCTION
Initiation of translation for cellular mRNAs can be broken down into three steps: the 40S ribosomal subunit, interacting with the initiation factor complex, binds capped polyadenylated mRNA at the 5′ end (step 1), scans the 5′ untranslated region (UTR) to reach the 5′ proximal AUG (step 2), is joined by the 60S subunit, and then initiates protein synthesis (step 3) (1). The recognition of the m7GpppG 5′ cap by the cap binding protein component (eIF4E) of eIF4F initiation complexes enhances ribosome recruitment to the mRNA. Viruses that replicate in the cytoplasm have evolved diverse mechanisms that deviate from canonical cap-dependent translation so as to circumvent host surveillance and overcome the need for RNA capping machineries that are typically confined to the nucleus (2–8). Cap-independent translation can be facilitated by an internal ribosome entry site (IRES), which varies greatly in size, structure, and mechanism depending on the virus (9). Despite their large diversity, IRESes perform the same function, i.e., assembly of the 40S ribosomal subunit in the vicinity of the correct initiation site, and skip the requirement for many, if not all, translation initiation factors that are typically involved in binding 40S subunits to cellular mRNAs (3, 10). When placed in the intergenic region of a discistronic mRNA, IRES elements are capable of driving translation of the downstream gene, independently of the first cistron. Translation can also be maintained in the context of monocistronic RNA molecules bearing 5′ barrier hairpin stem loops sufficiently strong to block entry of ribosomal complexes (11, 12). Here, we report on a unique mechanism of IRES-driven translation identified in an uncapped polyadenylated plant RNA virus, Triticum mosaic virus (TriMV), a member of the Potyviridae family.
IRES elements were first described for animal viruses in the Picornaviridae family (13–15). Picornavirus IRESes are embedded within relatively long 5′ UTRs of 600 to 1,200 bases and are characterized by stable RNA structures with high GC content and, frequently, multiple silent AUGs upstream of the authentic start-AUG initiation site (16). Depending on their structures, motifs, and modes of action, IRESes are classified as different types, with their preferred initiation sites specified through binding of a subset of translation initiation factors and cellular trans-acting factors (10). For several mammalian picornaviral IRESes, RNA-protein interactions promote ribosome complex loading onto a conserved Yn-Xm-AUG motif located hundreds of bases downstream of the 5′ end of the RNA, with “Y” representing a oligopyrimidine CU-tract of 8 to 10 nucleotides (nt), followed by a spacer “X” of about 10 to 20 unspecific nucleotides before an AUG codon (10, 17). Despite the presence of the multiple upstream AUG triplets within the 5′ UTR, the poliovirus (PV) “type I” IRES YX-AUG motif is silent, with the scanning ribosomal complex bypassing it to initiate translation at a further downstream AUG (18). In contrast, encephalomyocarditis virus (EMCV) “type II” IRESes position the correct AUG as part of the YX motif, and thus, translation starts at binding and without scanning (10, 17).
IRES-mediated internal initiation has now also been reported for several plant viruses, including some species in the Potyviridae (11, 19–22), Comoviridae (23), Tobamoviridae (24), Tombusviridae (25), and Luteoviridae (26) families. Plant virus IRESes are often AU-rich and are generally shorter and less structured than those of picornaviruses, and relatively little is yet known regarding their regulation and modes of action (5). Of those studied, members of the Potyviridae family bear a viral protein linked to the genome instead of a 5′ cap and are thus thought to be reliant on IRES-like elements for translation, with the prototypical element being the 143-nt-long 5′ leader of Tobacco etch virus (TEV; family Potyviridae) that contains two cap-independent regulatory elements (CIREs) of approximately 75 nt long each (19, 22, 27–29). CIREs drive internal initiation when placed in the intergenic region of a bicistronic RNA, consistent with IRES function. CIRE1 folds into an AU-rich pseudoknot structure with one loop that bears a 7-nt-long sequence, 5′-UACUUCU-3′, about 100 nt upstream of the initiation site. While the mechanism is not fully elucidated, this loop sequence exhibits complementarity to a conserved region of the 18S rRNA, a structural RNA component of the 40S ribosomal subunit, at nt 1131 to 1137 (28). Mutations within the 7-nt complementary sequence predicted to disrupt base complementarity resulted in an ∼80% decrease to translation in reporter assays (28), which is consistent with a hypothesis wherein the loop sequence directs recruitment of the 40S ribosomal subunit through base pairing. Similar 18S rRNA binding sites have been predicted in other plant viral cap-independent translation enhancers (7). However, to date these target sites remain poorly characterized.
Like TEV, the 5′ leader sequences of other potyviruses, including Turnip mosaic virus (TuMV) (20), Potato virus Y (PVY) (21), and Plumpox virus (PPV) (30), stimulate cap-independent translation. For example, the AU-rich 133-nt TuMV 5′ UTR can stimulate translation in both forward and reverse orientations, and the reverse sequence inhibits cap-dependent translation of cellular mRNAs in trans, presumably by competing for host translation factors (20). When a strong hairpin was placed at the very 5′ end of the mRNA, TuMV 5′ UTR-driven translation was ∼30% that observed without the stem-loop, a finding consistent with some level of internal initiation activity.
The diversity of potyviral translation enhancers is further emphasized by the recent discovery of a distinct species of the Potyviridae family, Triticum mosaic virus (TriMV; Poacevirus), with an exceptionally long (739-nt) 5′ UTR that contains 13 AUG triplets, with one of which (AUG13) is thought to serve as the primarily designated start codon (31, 32). The genome of TriMV is a polyadenylated positive-sense RNA of about 10,200 nt, has a 48.6-kDa viral protein linked to the genome (Vpg) at its 5′ end that replaces the 5′ cap, and encodes a single polyprotein (31, 32). Because of its low sequence homology to other potyviruses, TriMV was classified in a new poacevirus genus (31, 32). We have shown that the TriMV 5′ UTR facilitates translation at AUG13 in the context of an uncapped monocistronic mRNA (11). It does not require an open 5′ end for maximal translation. In fact, it retains full activity in the presence of a strong hairpin placed at the 5′ end of the monocistronic RNA, or when placed in the intergenic region of a dicistronic mRNA, revealing true IRES function (11). Our deletion analysis of the TriMV 5′ UTR delineated a 300-nt region sufficient to drive cap-independent translation in wheat germ extract in the context of an uncapped monocistronic mRNA and identified an 8-nt hairpin at nt 469 to 490 of the UTR critical for its translation function. Disruption of the stem structure abolished both cap-independent and IRES driven translation (11), correlating with a loss of binding to eIF4G, the scaffolding protein that promotes translation (33). We also demonstrated that TriMV translation is dependent upon the DEAD box eIF4A RNA helicase (33). Still undetermined is how the TriMV 5′ UTR specifically coordinates the assembly of the ribosomal complex and other translation factors proximal to AUG13.
Here, through assaying deletion mutations of the TriMV 5′ leader sequence, we demonstrate that the TriMV 5′ UTR encodes a core YX-AUG activator motif that is functionally equivalent to a picornavirus XY motif, exhibiting complementarity to 18S RNA and specifying the preferred site of translation initiation at AUG13. This element was necessary for the internal initiation function of the TriMV 5′ UTR both in vitro and in vivo. Phylogenetic analysis of members of the Potyviridae family and functional comparisons between the TriMV 5′ UTR and that of the Bellflower veinal mottle virus (BVMoV), a newly discovered potyvirus of bellflower (34), suggest a possible remnant motif among potyviruses. To our knowledge, this is the first evidence of a IRES-driven motif-based mechanism for translation conserved in both plant- and animal-hosted picornaviruses.
RESULTS
Identification of two distinct TriMV 5′ UTR sequences required for optimal IRES activity.
We previously established that the 739-nt TriMV 5′ UTR is sufficient to drive cap-independent translation of uncapped monocistronic mRNA and acted as an IRES when placed in the intergenic region of a standard bicistronic construct (11). By deleting regions of the TriMV leader sequence, we demonstrated that the final 300 nt of the TriMV 5′ UTR (spanning nt 442 to 709 of the 739-nt sequence) are sufficient to drive cap-independent translation in the context of the uncapped monocistronic mRNA, at least in wheat germ extract (11).
To better define the portions of this sequence relevant to IRES function, we endeavored to measure the effects of these and additional TriMV 5′ UTR sequences when inserted between renilla and firefly luciferase reporter genes in the context of a bicistronic RNA IRES reporter construct (Fig. 1A). Expression of the second cistron (firefly luciferase) depends on internal initiation, while the expression level of the first cistron (renilla luciferase) is driven by a 5′ cap. Bicistronic mRNAs were in vitro synthesized, capped, polyadenylated, and translated in wheat germ extract prior to calculating the ratio of firefly luciferase to renilla luciferase light units, with measurements greater than the ratio observed for a “no insert” control defined as representative of IRES activity.
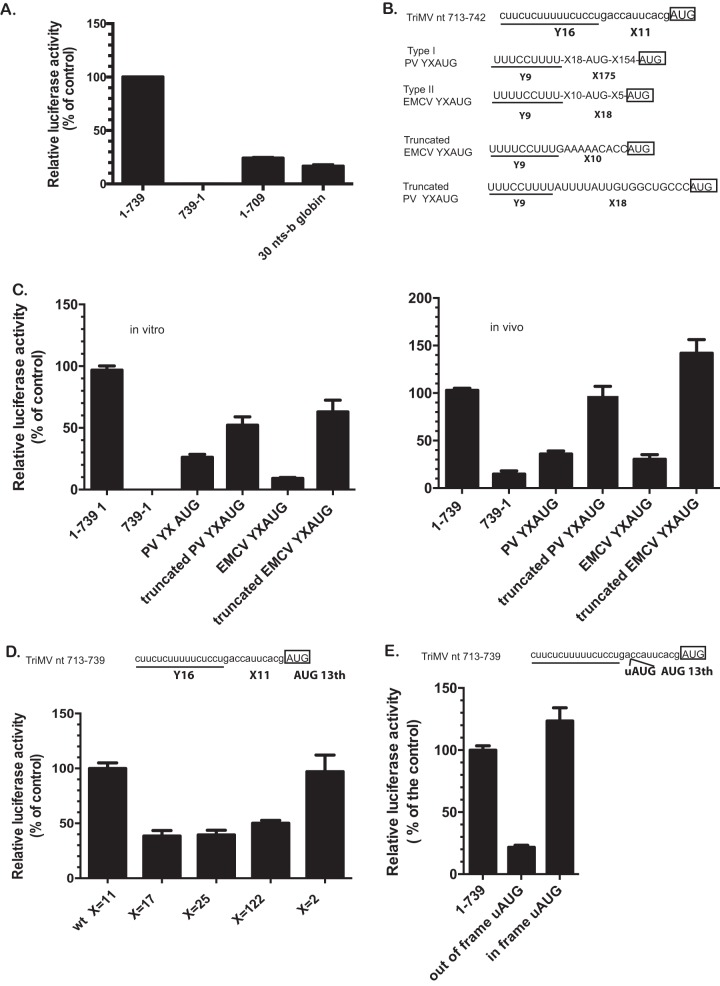
FIG 1.
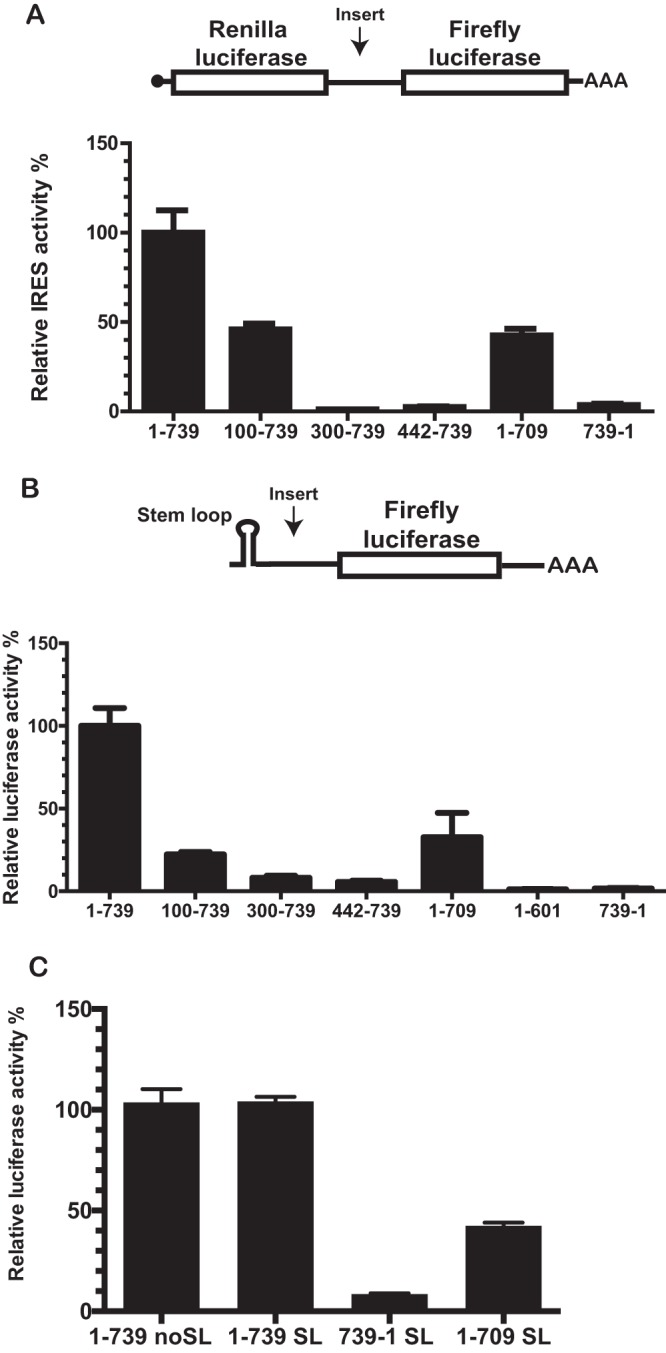
The 5′ and 3′ borders of the TriMV 5′ UTR are necessary for the TriMV IRES activity. (A) Schematic diagram of the bicistronic dual-luciferase reporter RNA and the positions of the insertions of the RNA element tested. Translation of the renilla luciferase gene is cap mediated, but translation of the downstream firefly luciferase gene can be directed only by internal initiation driven by the RNA sequence inserted into the intergenic region. The internal initiation ability is quantified as the ratio of firefly luciferase to renilla luciferase activities. The ratio of firefly to renilla luciferase activities in wheat germ extract of the bicistronic mRNAs was determined by a m7 GpppG cap at the 5′ end and the TriMV 5′ UTR (construct 1-739) or the nonfunctional TriMV reverse complementary sequence (construct 739-1) or the deletion mutants in the intergenic region. (B) Schematic diagram of monocistronic firefly luciferase reporter RNA containing a stable hairpin insertion (ΔG = –34 kcal) immediately at the 5′ end of the mRNA (12). The relative luciferase activities in wheat germ extract of the TriMV 5′ UTR deletion mutants with the strong hairpin is shown and is relativized to that of the TriMV wild-type sequence. (C) Relative luciferase activity in oat protoplasts of the reporter mRNAs containing the full-length TriMV leader (construct 1-739 SL) or the deletion mutant missing the last 30 nt (construct 1-709 SL) as 5′ UTR with the strong hairpin, normalized to values for a m7GpppG-capped polyadenylated renilla mRNA used as an internal control, which we coelectroporated at a 1:10 ratio. As a control, we included the full-length TriMV leader sequence as 5′ UTR in the absence of the strong stem-loop (construct 1-739 noSL) and the nonfunctional TriMV reverse complementary sequence (construct 739-1 SL).
We first compared IRES activity for a defined set of informative deletion mutant constructs (nt 100 to 739, nt 300 to 739, nt 442 to 739, and nt 1 to 709) to that of the full-length UTR (construct 1-739) and the nonfunctional reverse complementary sequence of the viral 5′ UTR (construct 739-1) as a negative control (Fig. 1A). The entire TriMV 5′ UTR was required for optimal IRES-driven translation in this assay, with deletion of nt 1 to 99 (construct 100-739) sufficient to significantly reduce translation (45% relative to full-length 5′ UTR). Further deletions from the 5′ end (constructs 300-739 and 442-739) reduced translation to background levels, as did the reverse complement (construct 739-1). Similar effects were observed in the context of a monocistronic luciferase reporter RNA bearing a 5′ barrier hairpin stem-loop sufficiently strong (ΔG = –34 kcal) to block 5′ entry of ribosomal complexes and thereby requiring internal initiation for reporter gene expression (11, 12) (Fig. 1B).
Surprisingly, a deletion of the 30 nt upstream of the firefly luciferase start AUG (construct 1-709) significantly impacted translation in both dicistronic and monocistronic systems (Fig. 1A and B) (∼40% relative to full-length 5′ UTR), prompting us to investigate the relevance of these deleted sequence also in oat protoplast cells, a natural host of the virus (Fig. 1C). As anticipated, the full-length TriMV 5′ UTR was capable of driving cap-independent translation in the absence (construct 1-739) or the presence of the 5′ barrier hairpin (construct 1-739 SL) with similar efficiency in protoplasts and, again, the deletion of the 30 nt upstream of the AUG13 impaired translation of the TriMV SL-mRNA down to 50% of its activity in this system.
Identification of a novel YX-AUG translation initiation motif in the TriMV IRES.
To assess whether the 3′ 30-nt region encoded an important signaling motif (as opposed to serving as a nucleotide spacer), we first tested the effects of replacing the region with an unrelated 30-nt sequence derived from human β-globin RNA (Fig. 2A). This modification yielded reduced IRES activity, similar to deleting those bases (compare to construct 1-709 in Fig. 1), suggesting the presence of a cis-acting functional element. Subsequent sequence analysis of the native 30-nt region revealed a 16-nt polypyrimidine tract, 5′-CUUCUCUUUUUCUCCU at nt 713 to 728, 11 nt upstream of the designated AUG13 (Fig. 2B). Interestingly, this sequence pattern was remarkably similar to known Yn-Xm-AUG motifs (Yn being a polypyrimidine CU-tract separated by an Xm spacer with a nonspecific nucleotide sequence from the preferred AUG codon) previously described for a subset of animal picornavirus IRESes and involved in defining the recognition of the correct initiation site in the presence of multiple AUG triplets (10, 17). The two best-characterized YX motifs are the type I poliovirus (PV) motif, Y9X18-aAUG-X154-bAUG, and the type II EMCV motif Y9X10-aAUG-X5-bAUG (Fig. 2B) with a cryptic aAUG triplet out of frame of the downstream favored bAUG (18, 35). An important feature of YX-AUG motifs in picornaviruses is the length of the X spacer between the CU-tract and the preferred AUG, which are 18 and 175 nt, respectively, for the EMCV and the PV motifs (18, 35). The spacer determines the scanning ability of the ribosomal complex to find the preferred start codon.
FIG 2.
The functional AUG is part of a functional equivalent of the picornavirus YX-AUG motif. (A) The relative luciferase activity in wheat germ extract of the TriMV 5′ UTR with the deletion of the last 30 nucleotides (1-709) or with the last 30 nt replaced with unrelated human β-globin sequence (30-nt b-globin) is relativized to that of the TriMV wild-type sequence in the presence of the strong hairpin. (B) The TriMV 5′ UTR sequence (GenBank accession number FJ669487) bears an Y16X11-AUG like motif at positions nt 710 to 739, upstream of the correct AUG. The Y is a 16-nt polypyrimidine tract. X is the spacer sequence with 11 random nucleotides. These motifs are commonly found in picornavirus IRESes, exemplified with the motifs in PV type I IRES Y9X18-aAUG-X154-bAUG, and in the EMCV type II IRES Y9X10-aAUG-X5-bAUG (10). The motifs contain a cryptic aAUG triplet, which is out of frame of the downstream correct bAUG. The truncated motifs are missing all the sequences downstream of the cryptic AUG. The AUG of the luciferase corresponds to the boxed AUG codon. (C) The relative luciferase activity in wheat germ extract (on the left) and in oat protoplasts (on the right) of the chimeric TriMV 5′ UTRs with the last 30 nt being replaced with the mammalian YX-AUG motif counterparts is relativized to that of the TriMV wild-type sequence in the SL-mRNA reporter. For the oat protoplast assays, the reporter mRNAs were coelectroporated with a m7GpppG capped polyadenylated renilla mRNA used as an internal control at a 1:10 ratio. (D) The relative luciferase activity in wheat germ extract of the TriMV 5′ UTR sequence with an extended (X17, X25, and X122) or reduced (X2) spacer length is relativized to that of the TriMV wild-type sequence, in the presence of the strong hairpin. (E) The relative luciferase activity in wheat germ extract of the TriMV 5′ UTR sequence with an upstream AUG inserted 2 bases downstream of the CU-tract at position nt 727 and placed out of frame or in frame with the downstream luciferase AUG.
To determine whether TriMV encodes a functional YX motif similar to EMCV and PV, we first tested the effects of replacing the TriMV 30-nt-sequence region with select mammalian virus YX counterparts (Fig. 2C). In these chimeric constructs, the initiation codon of the luciferase SL-mRNA reporter corresponded to the second bAUG triplet in each of the motifs. Both native picornavirus YX AUG motifs (PV YXAUG and EMCV YXAUG) supported TriMV IRES activity both in wheat germ extract and in oat protoplast assays, albeit relatively poorly (compare Fig. 2A and C). In contrast, truncated versions engineered to place the luciferase initiation site at the position of the first aAUG triplet by reducing the distance between the CU-track and the AUG down to 10 and 18 nt, respectively, for the EMCV and the PV motifs and also removing the presence of an out-of-frame AUG triplet within the X spacer sequence supported TriMV IRES translation above 50% of the wild-type level in wheat germ extract (Fig. 2C, left) and completely in protoplasts (Fig. 2C, right). This result reveals that TriMV encodes a functional YX-AUG motif with activities homologous to its mammalian virus counterparts.
These observations led us to next evaluate the importance of the TriMV “X” region’s spacer length in the context of a putative YX-AUG model regulatory element. To this end, we increased spacer length from X11 nucleotides to X17, X25, and X112 with 6-, 14-, and 102-nt insertions, respectively (Fig. 2D), with random sequences that did not contain AUGs or stop codons and predicted by MFOLD to be unlikely to cause changes to the overall TriMV 5′ UTR conformation. As shown in Fig. 2D, a 6-nt increase in spacer length from 11 to 122 nt reduced translation to 60% relative to the wild-type sequence. In contrast, shortening the spacer length down to 2 nt (X2) had no effect, ergo a short X spacer seems vital to the integrity of TriMV YX-AUG motif function.
We also reasoned that, should the element be a functional YX motif, the first AUG following the CU-tract, and within a short distance, should be preferentially recognized by the ribosomal complex. To test this hypothesis, we inserted an upstream AUG (uAUG) codon within the spacer sequence 2 nt downstream of the CU-tract, out of frame with start AUG of the downstream luciferase reporter sequence (Fig. 2E). Translation, were it to be initiated at the uAUG, would shift frames, thereby abolishing translation of the luciferase reporter. Consistent with our hypothesis, the out-of-frame AUG reduced luciferase levels to ∼20% of wild type (Fig. 2E). In contrast, when the uAUG was placed in frame with the luciferase initiation site, translation was fully restored.
Together, these genetic analyses indicated that the TriMV 5′ UTR encodes a bona fide YX-AUG motif, similar to and functionally redundant with YX motifs previously described for animal picornaviruses. Moreover, these data support a model wherein the preferred initiation site of the TriMV 5′ UTR is embedded within the YX-AUG motif (AUG13) at requisite short distance from the CU-rich tract, which is perhaps consistent with overcoming limitations to the scanning ability of ribosomal complexes.
The YX-AUG CU-tract likely functions as a target site for direct 18S rRNA binding.
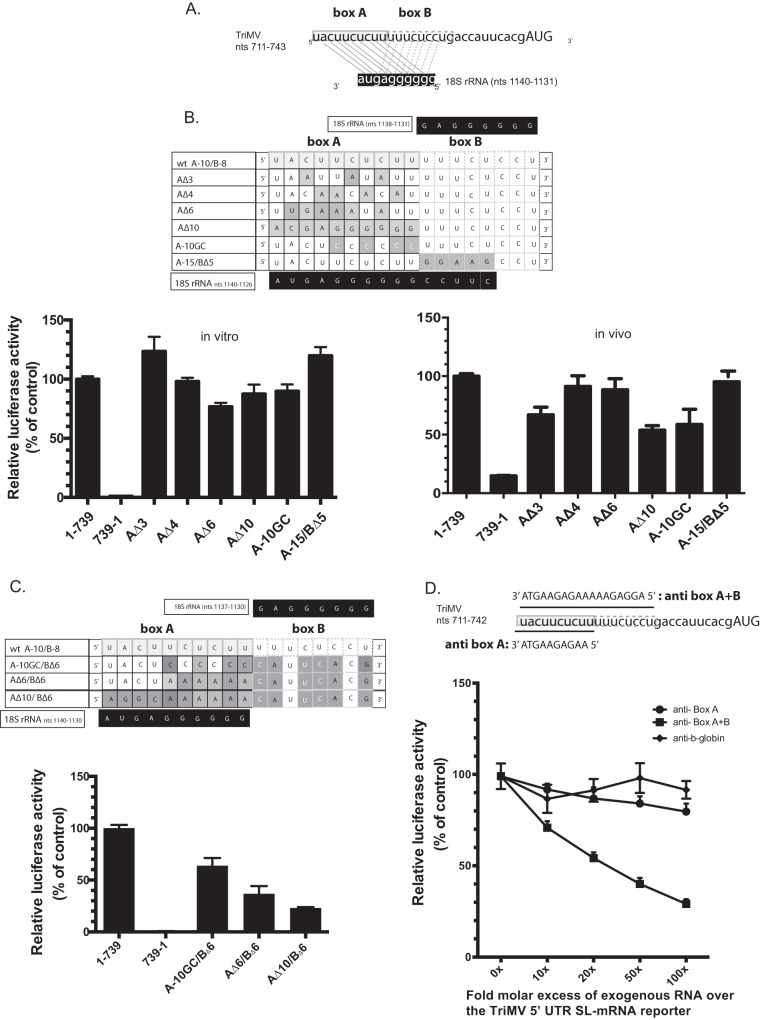
The results presented above prompted us to explore whether the TriMV YX-AUG motif might function as a ribosomal targeting site. Interestingly, sequence analysis of the motif and its immediate upstream nucleotides revealed two adjacent sites (dubbed “box A” and “box B”), both exhibiting complementarity to a G-rich tract within a highly conserved region of the 18S rRNA (Fig. 3A). The relevant G-rich tract of the 18S rRNA is found within a previously defined and conserved rRNA’s “active” domain (nt 812 to 1233), involved in interactions with other mRNA regulatory elements (36). The TriMV box A is a 10-nt sequence spanning TriMV nt 711 to 720 (5′-UACUUCUCUU-3′) and shares complementarity to the 18S rRNA sequence 5′-GGGGGGAGUA-3′, encoding the same 7-nt sequence found in the Tobacco etch virus IRES element 5′-UACUUCU-3′ necessary for translation and similarly proposed to bind the same wheat 18S rRNA region (28). Box B is an 8-nt sequence 5′-UUUCUCCU-3′ that can base pair to the same sequence of the 18S rRNA 5′-GGGGGGAG-3′ (Fig. 3A).
FIG 3.
The TriMV YX-AUG motif likely functions as target sites for the 18S rRNA binding (A) Sequence showing the two adjacent putative binding sites (box A and box B) of the TriMV IRES at positions nt 711 to 720 and nt 721 to 728 to a highly conserved region of the 18S rRNA at positions nt 1123 to 1140. (B) The mutated bases within the TriMV box A 18S rRNA target site that either partially or fully decreased (AΔ3, AΔ4, AΔ6, and AΔ10), increased (A-15/BΔ5) base pairing interaction or strengthened (A10-GC with the wobble base pairs within box A replaced with GC pairs) are shaded. The wild-type sequence is annotated as “wt A-10/B-8”. The relative luciferase activity in wheat germ extract (on the left) and in oat protoplasts (on the right) of the different TriMV mutants is relativized to that of the TriMV wild-type sequence. For the oat protoplast assays, the reporter mRNAs were coelectroporated with a m7GpppG capped polyadenylated renilla mRNA used as an internal control at a 1:10 ratio. (C) The mutated bases within the TriMV box A and box B 18S rRNA target sites that reduced the overall base pairing interaction and the CU-richness of the region are shaded. The wild-type sequence is annotated as “wt A-10/B-8”. The relative luciferase activity of the TriMV 5′ UTR with different mutations across the box A and box B 18S rRNA binding sites in wheat germ extract is relativized to that of the wild-type sequence in the presence of the strong hairpin. (D). trans-Inhibition assay of the TriMV 5′ UTR SL-mRNA with increasing molecular excess of antisense single-stranded DNA oligonucleotides targeting the box A (anti-BoxA) or both box A and box B (anti-BoxB) 18S rRNA binding sites in wheat germ extract. As a control DNA oligonucleotides targeting unrelated human β-globin sequence was added. A 0- to 100-fold molar excess of the antisense oligonucleotides was added to the reaction.
Mutations designed to partially or fully disrupt the putative box A 18S binding site while maintaining the box B sequence (AΔ3, AΔ4, AΔ6, and AΔ10) or to strengthen putative box A-18S interactions by replacing the wobble GU base pairing into GC (A-10GC) only moderately affected IRES activity in vitro (Fig. 3B, left) and had a slightly more pronounced effect in oat protoplasts (Fig. 3B, right). Moreover, extension of box A from 10 to 15 bases had no net effect on translation in vitro or in vivo (A-15/BΔ5, Fig. 3B). However, a combination of box A and box B mutations that disrupted overall complementarity to 18S rRNA and the CU-richness of the region reduced IRES activity by as much as 80% (Fig. 3C). These results are at consistent with there being redundancy in function for the two cis-acting oligopyrimidine boxes.
Next, we attempted to directly interfere with potential box A/B-18S base pairing interactions using antisense single-stranded DNA oligonucleotides, in virtue of their sequence complementarity in vitro (Fig. 3E). To this end, we added DNA oligonucleotides consisting of sequences complementary to the box A sequence alone (anti-BoxA), both box A and box B sequences (anti-BoxA+B), or the human β-globin as a negative control (anti-b-globin), up to a 100-fold molar excess relative to the TriMV 5′ UTR SL-mRNA in wheat germ extract. We normalized translation levels to the no antisense DNA oligonucleotide added control, defined as 100% translation. As anticipated, the unspecific DNA oligonucleotides (anti-b-globin) had no effect on translation, whereas, consistent with a role for the box A/B sequence in regulating one or more trans-mediated effects, the TriMV 5′ UTR SL-mRNA reporter steadily decreased in the presence of an increasing fold molar excess of the antisense DNA oligonucleotide targeting both box A and box B 18S binding sites (Fig. 3E). Loss of translation was not significant for the anti-BoxA treatment alone, a finding consistent with our genetic result, wherein both box A and box B had to be mutated in order to generate a loss-of-function phenotype (Fig. 3D).
Taken together, the data are consistent with a model wherein the TriMV YX-AUG motif and its two oligopyrimidine boxes function as target sites for 18S rRNA binding, similar to the mechanism previously proposed for the prototype potyviral TEV IRES for the translation at its single AUG located about 100 nt downstream of the motif (28).
Unrelated viral sequences that bear putative 18S binding sites can substitute for the TriMV YX-AUG motif function.
To further characterize the relevance of the 18S rRNA targeting site in the TriMV IRES activity, we swapped the last 30 nt of the UTR with putative 18S binding sites derived from translation enhancer elements of unrelated uncapped plant viral RNAs (Fig. 4). These sites show complementarity to various stretches within the 3′ end region of the 18S rRNA and included (i) the 17-nt conserved sequence (GGAUCCUGGGAAACAGG) within the Barley yellow dwarf virus-like cap-independent translation element (BTE) present in some members of the Luteoviridae and Tombusviridae families (37), (ii) a 19-nt sequence from an 100-nt internal region element upstream of the coat protein gene of Hibiscus chlorotic ringspot virus (HCRV; Carmovirus) (CUGAUUAAGGCUGAAUCAG) (38), and (iii) a 47-nt CU-rich sequence from the Blackcurrant reversion virus (BRV; Comoviridae) RNA 2 leader sequence (23). The BTE normally drives translation at the 5′-end proximal AUG from its 3′-end position in the uncapped, nonadenylated viral genome with the 17-nt sequence, including a predicted 6-nt “GAUCCU” 18S binding site (8). The 100-nt region (nt 2394 to 2566) upstream of the coat protein gene within the HCRV genomic RNA drives internal initiation of the P38 viral coat protein (38). Sequences but not the secondary structure within a 19-nt region at nt 2494 to 2512 were identified important for its activity (38). These included a 6-nt CUGAUU putative 18S binding site (38). The BRV leader sequence was reported to drive internal initiation at its single AUG, with the selected region UGCUCUUUUUCCUUCCUUCCUUCGCUUUUGUUUUGGUUUGCUUUACC containing two of the putative six 18S rRNA motifs within the 161-nt BRV uncapped 5′ UTR, complementary to the 18S rRNA from bp 1113 to 1123 (23). It is worth noting that while important for translation in their natural viral context, these selected viral regions are not reported to exert translational activity on their own. When tested in the context of our TriMV YX-AUG swap assay (Fig. 4A); however, the non-CU-rich HCRV and the BRV sequences with their putative 18S binding sites supported translation up to 70% of the wild-type level, although the BTE element had barely any stimulatory effect, similar to that conferred by the 30-nt b-globin negative-control sequence. To validate the relevance of the 18S rRNA binding sites in TriMV translation, we introduced mutations designed to disrupt the putative 6-nt sequence complementary to the 18S rRNA within the 19-nt HCRV sequence (CUGAUU into GGAUCC construct +19nts mutant HCRV) in the chimeric TriMV 5′ UTR. The mutations resulted in a drastic loss of translation (Fig. 4B). Moreover, when we directly interfered with the potential HCRV-18S rRNA base pairing interactions using antisense oligonucleotides to the 19-nt HCRV sequence (anti-HCRV), translation of the chimeric TriMV RNA steadily decreased (Fig. 4C). As anticipated, the unspecific DNA oligonucleotides (anti-β-globin) and the oligonucleotides targeting the native TriMV box A-box B region had no effect on translation (Fig. 4C). Altogether, these data are in line with a model wherein 18S rRNA targeting may play a key role in TriMV YX-AUG motif activity.
FIG 4.
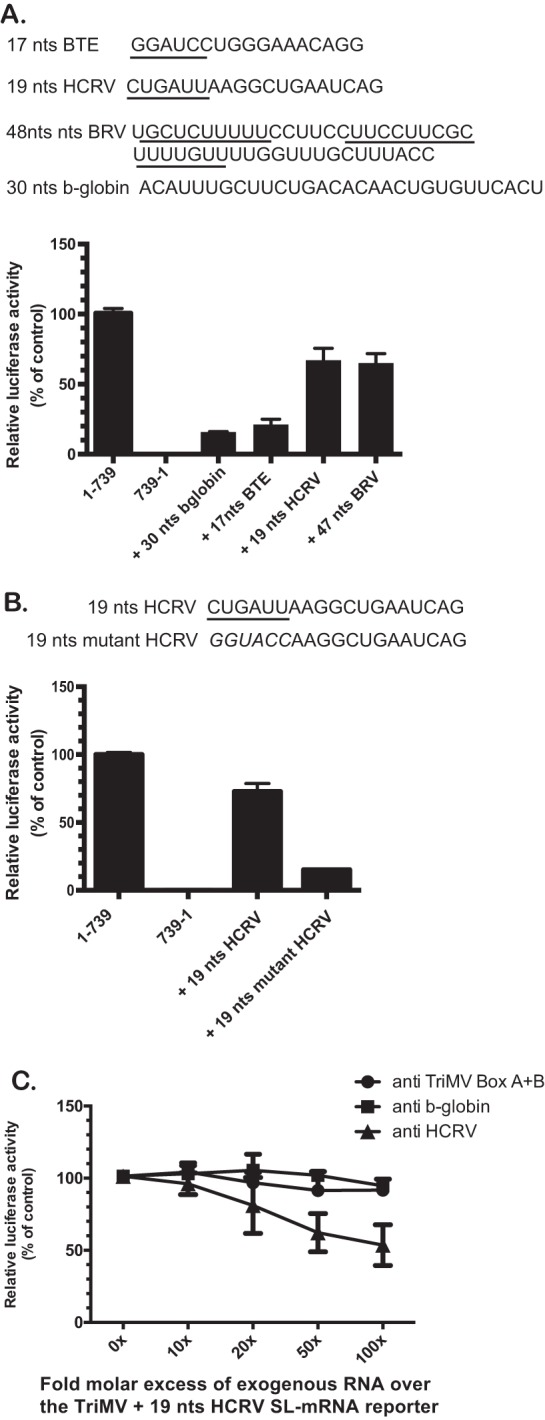
The TriMV YX-AUG motif can be substituted with putative 18S rRNA binding sites from unrelated viruses. The sequence of the viral motifs used to swap the last 30 nt of the TriMV 5′ UTR is shown. Sequences are from Blackcurrant reversion virus (BRV) RNA 2 (accession no. NC_003502.1; nt 115 to 161), Hibiscus chlorotic ringspot virus (HCRV; accession no. X86448; nt 2494 to 2512), and Barley yellow dwarf virus (accession no. NC_004750.1; nt 4837 to 4853). (A) The relative luciferase activity in wheat germ extract of the chimeric TriMV 5′ UTRs with the last 30 nt being replaced with unrelated viral sequence with reported putative 18S binding sites is relativized to that of the TriMV wild-type sequence in the SL-mRNA reporter. (B) The relative luciferase activity in wheat germ extract of the chimeric TriMV 5′ UTRs with the last 30 nt replaced with the 19-nt HCRV sequence (+19nt HCRV) or a modified 19-nt HCRV sequence (+19nt mutated HCRV) with mutations that disrupted the putative 6-nt complementary sequence to the 18S rRNA is relativized to that of the TriMV wild-type sequence in the SL-mRNA reporter. (C) trans-Inhibition assay of the chimeric TriMV+19nt HCRV SL-mRNA with increasing molecular excess of antisense single-stranded DNA oligonucleotides targeting the 19-nt HCRV sequence (anti-HCRV) in wheat germ extract. As controls, DNA oligonucleotides targeting unrelated human β-globin sequence and the native TriMV box A + box B were added. A 0- to 100-fold molar excess of the antisense oligonucleotides was added to the reaction.
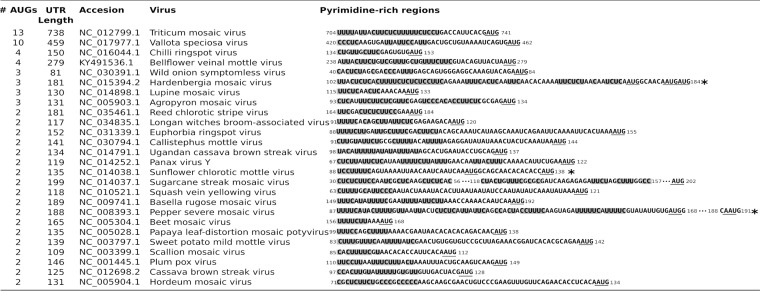
YX-AUG motifs conserved features of potyviruses bearing multi-AUG 5′ UTRs.
Interestingly, additional members of the Potyviridae family encode 5′ UTRs bearing multiple AUGs, with sequence analysis of 153 potyviral 5′ UTRs identifying 35 multi-AUG 5′ UTRs, 8 of them bearing more than two AUG codons (Fig. 5). To determine whether YX-AUG motifs represent a conserved regulatory feature of this class of viruses, we aligned the 35 potyviral 5′ UTRs that bear more than one AUG in their predicted 5′ UTRs and searched for CU-rich tract signatures upstream of the putative initiation site (Fig. 5). A total of 26 multi-AUG 5′ UTRs, which represent 16.9% of potyviruses, encode a CU-rich tract upstream of the last AUG triplet. Three potyviral 5′ UTRs showed a polypyrimidine tract upstream of the first AUG codon (indicated by an asterisk in Fig. 5). This analysis suggested that the CU-rich region and/or the YX-AUG like motif might be broadly conserved within the Potyviridae family.
FIG 5.
Sequence alignment of different potyvirus 5′ UTRs bearing multiple AUGs. Highlighted in gray shades are the CU-rich tract identified to close proximity of the last AUG codon. Putative motifs found closest to the first AUG codon are indicated by asterisks. Sequence symbols: *, putative motif found in the first AUG; …, stretch of non-polypyrimidine-rich sequence.
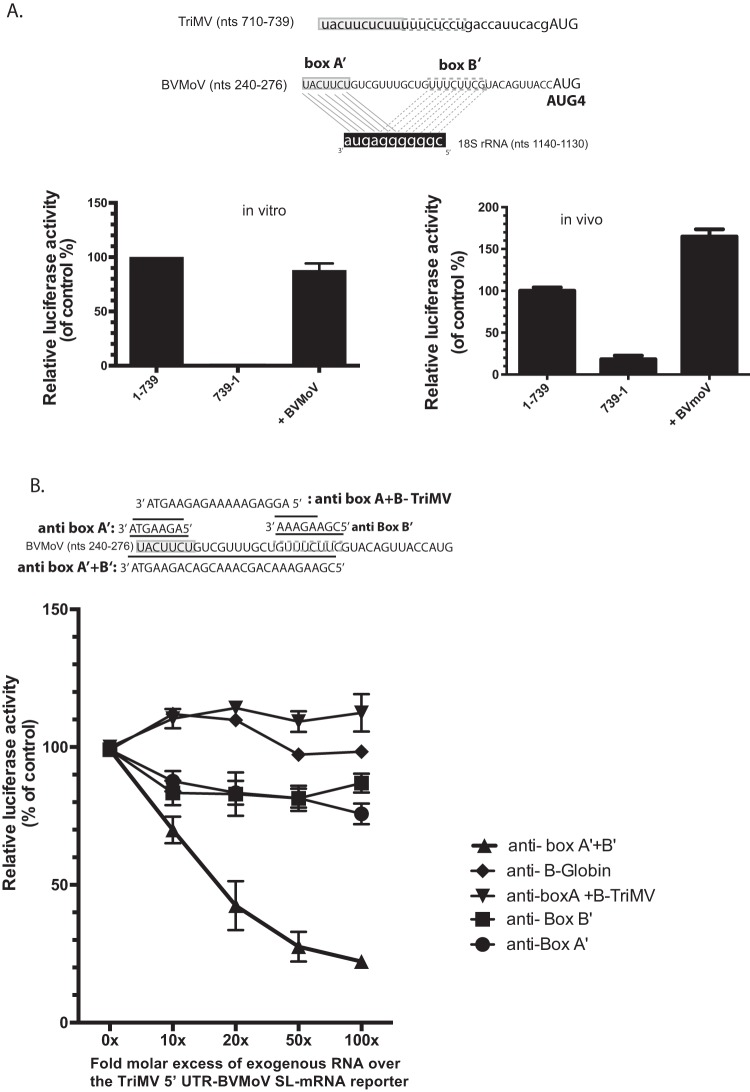
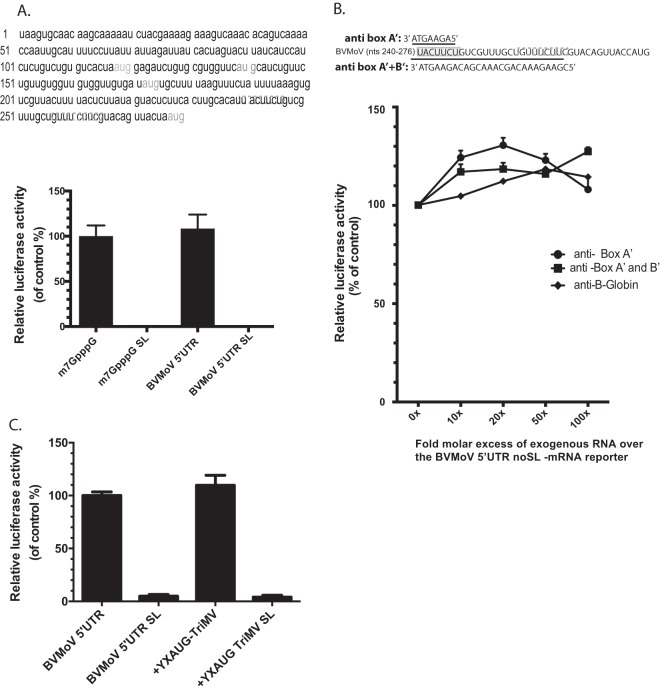
Although determining the functional relevance of diverse potyviral YX-AUG-like motifs was beyond the scope of this study, we did become particularly interested in one of these potyviruses, the recently emerged bellflower pathogen, Bellflower veinal mottle virus (BVMoV) that has a 276-nt uncapped leader sequence with four AUGs (39). Analysis of the BVMoV 5′ UTR sequence revealed a 35-base CU-rich region just upstream of the AUG4 codon (Fig. 6), with this region encoding two uncoupled putative 18S rRNA binding sites, box A′ (5′ UACUUCU 3′) and box B′ (5′ UUUCUUCG 3′), 11 nt apart from one another, and with some overlapping sequences to that of the TriMV 18S rRNA target sites and complementary to the same region of the 18S rRNA (Fig. 6A). The CU-rich track was separated from the AUG codon by 12 bases.
FIG 6.
A 35-nt sequence from the BVMoV leader sequence can functionally substitute for the TriMV YX-AUG function in translation. (A) Boxed are the two uncoupled putative box A′ and box B′ binding sites within the BVMoV 5′ UTR sequence at position nt 240 to 247 and nt 258 to 265 (accession number KY491536.1) upstream of the fourth AUG. The relative luciferase activity of the TriMV 5′ UTR with the last 30 nt swapped with a 35-nt sequence of the BVMoV in wheat germ extract (on the left) and in oat protoplasts (on the right) is relativized to that of the TriMV wild-type sequence in the presence of the strong hairpin. For the oat protoplast assays, the reporter mRNAs were coelectroporated with a m7GpppG-capped polyadenylated renilla mRNA used as an internal control at a 1:10 ratio (B). trans-Inhibition assay of the chimeric TriMV 5′ UTR-BVMoV SL-mRNA construct with increasing molecular excess of antisense single-stranded DNA oligonucleotides targeting the box A′ alone (anti-BoxA′), box B’ (anti-BoxB′) alone, both box A′ and box B′ (anti-BoxB′), and box A and box B of the native TriMV (anti-BoxA+BoxB-TriMV) 18S rRNA binding sites in wheat germ extract. As a control, DNA oligonucleotides targeting unrelated human β-globin sequence were added. A 0- to 100-fold molar excess of the antisense oligonucleotides was added to the reaction.
Taking advantage of the reliance on an YX-AUG motif for the TriMV IRES activity, we swapped the last 30 bases of the TriMV 5′ UTR with the 35-nt sequence of the BVMoV leader sequence to test whether functionality in IRES activity was transferable and found it to drive 90% translation relative to the native TriMV motif in wheat germ extract (Fig. 6A, left), and with an even greater stimulatory effect observed in oat protoplasts (Fig. 6A, right). Similar to the TriMV box A/B sequence (Fig. 3D), increasing the fold molar excess of antisense DNA oligonucleotides targeting both BVMoV box A′ and box B′ steadily decreased the translation of our chimeric TriMV-BVMoV 5′ UTR SL-mRNA (Fig. 6B), with translation only moderately affected by oligonucleotides targeting box A′ or box B′ alone. Antisense DNA oligonucleotides targeting the native TriMV 18S rRNA binding sites (anti-BoxA+B TriMV) had no effect on the translation of the chimeric TriMV with the BVMoV A/B boxes. Combined, these results confirmed a functional equivalence for BVMoV and TriMV YX-AUG-like motifs.
To examine whether the BVMoV YX-AUG motif was also functional in its native sequence context, we first tested the translation activity of the full, 276-nt BVMoV 5′ UTR in the context of an uncapped polyadenylated monocistronic reporter RNA with the luciferase initiation site corresponding to the AUG4 codon of the BVMoV 5′ UTR (Fig. 7). The uncapped BVMoV 5′ UTR was capable of driving translation with as great efficiency as a m7GpppG-capped polyadenylated RNA control in wheat germ extract, a finding in line with a cap-independent translation function (Fig. 7A). However, the presence of a 5′ stem-loop structure (BVMoV-SL, m7GpppG SL) abolished both cap-dependent and BVMoV 5′ UTR-driven translation, demonstrating its lack of IRES activity (Fig. 6A). Antisense oligonucleotides also had no effect on BVMoV 5′ UTR translation (Fig. 6B), so that the BVMoV YX-AUG-like motif’s activity as a potential target for 18S rRNA, observed in the context of the TriMV 5′ UTR (Fig. 5), is apparently suppressed or secondary to other enhancer elements in the context of the full-length BVMoV 5′ regulatory sequence. Swapping the last 36 bases of the BVMoV 5′ UTR with the 30-nt YX-AUG sequence of the TriMV leader sequence maintained the BVMoV 5′ UTR-mediated translation, which is in line with an interchangeable character of the motifs. However, it was not sufficient to drive IRES activity of the chimeric BVMoV SL mRNA (Fig. 6C). In sum, although YX-AUG-like motifs appear to be a broadly conserved feature of potyvirus 5′ UTRs with multiple AUGs, their relative contributions to translation initiation may be variable depending on the viral species and, possibly, their strategy of translation.
FIG 7.
The BVMoV 5′ UTR-driven translation may not be dependent of the YX-AUG like motif. (A) Sequence of the 276-nt BVMoV 5′ UTR (accession number KY491536.1). In gray are the multiple AUGs. Boxed are the two putative 18S rRNA binding sites. The BVMoV YX-AUG like motif covers region nt 242 to 275. (A). The relative luciferase activities in wheat germ extract of the BVMoV 5′ UTR reporter mRNA with (BVYM 5′ UTR SL) or without (BVMoV 5′ UTR) the strong hairpin are relativized to that of an m7GpppG-capped polyadenylated mRNA control with vector sequence. (B) trans-Inhibition assay of the BVMoV 5′ UTR mRNA construct with increasing molecular excess of antisense single-stranded DNA oligonucleotides targeting the box A′ alone (anti-BoxA′), both box A′ and box B′ (anti-BoxB′) 18S rRNA binding sites in wheat germ extract. As a control, DNA oligonucleotides targeting unrelated human β-globin sequence were added. A 0- to 100-fold molar excess of the antisense oligonucleotides was added to the reaction. (C) The relative luciferase activities in wheat germ extract of the BVMoV 5′ UTR reporter mRNAs with the last 36 nt swapped with the 30-nt TriMV YX-AUG motif with or without the strong hairpin are relativized to that of the native BVMoV 5′ UTR reporter mRNA.
DISCUSSION
Despite a rich diversity of sequences and structures, viral IRES elements perform similar functions in diverse cell systems: recruiting ribosomal complexes to the functional AUG so that they initiate at the correct start codon without having to scan through complex 5′ UTR regulatory secondary structures (1). IRESes can recruit the ribosomal complex via the formation of highly structured RNAs, using cellular trans-acting factors, or by base pairing to the 18S rRNA (9). We have previously shown that Triticum mosaic virus (TriMV) devotes at least 7% of its genome, which corresponds to the entire 739-nt 5′ UTR, to internally directing ribosomes to the correct AUG (11) (Fig. 1). Here, we identify and characterize a key 30-nt region of this UTR that plays a key role in defining this preferred initiation site.
Unlike most reported plant virus IRES elements (2, 5, 6), the TriMV IRES bears 13 AUG triplets. We previously showed that translation is primarily initiated at AUG13 (11). It is clear that such translation differs from the classic scanning mechanism where the ribosomal complex enters from the 5′ end of the mRNA and scans in a 5′-to-3′ direction for the correct 5′-proximal initiation site (1). Leaky scanning has been proposed for the translation of the correct AUG in Plum pox virus (PPV) RNA, another member of the Potyviridae family (30). This 146-nt potyviral 5′ UTR contains an in-frame upstream AUG at positions nt 36 to 38 preceding the authentic initiator AUG at positions nt 147 to 149 (30). When the upstream AUG was placed downstream of a strong Kozak consensus sequence, it became the preferred initiation site, supporting a leaky scanning mechanism even in the absence of a 5′ cap. In our case, translation at AUG13 for the TriMV 5′ UTR is regulated by the ability of RNA elements to drive internal translation (11), and we elucidate here how TriMV RNA coordinates this event using a cis-acting YnXm-AUG-like motif functionally interchangeable with prototype YX-AUG motifs found in animal picornaviral IRESes (Fig. 2) and exposed also in other members of the Potyviridae (Fig. 5 and 7).
Mechanistically, in picornaviruses, the position of the polypyrimidine CU-tract (Yn) determines the landing site of the ribosomal complex for the correct AUG and contributes to internal initiation (10, 17). For the EMCV type II-like IRES, translation initiates at the AUG embedded within the motif, which is 18 nt downstream of the polypyrimidine tract (10, 17, 35). The EMCV IRES harbors 12 AUGs, with the 11th AUG being the preferred initiation site. In fact, the polypyrimidine CU-tract within the EMCV IRES is followed by three consecutive AUGs. Alteration of the length of the Xm spacer separating the CU-tract and each of the AUG results in differential preference for initiation between the 10th, 11th, and 12th AUGs (35). If the Xm sequence is shortened, translation favorably occurs at AUG12, which is positioned 12 nt downstream of AUG11. If the Xm sequence is extended, AUG10 is recognized as the initiation site (35). For other picornaviral serotypes, including poliovirus, ribosomal complexes bypass the AUG triplet embedded within the YX-AUG motif and scan to initiate translation at a downstream AUG (18). Thus, not all YX-AUG motifs are created equal in terms of form or function.
Here, we report the first example of a plant virus XY-AUG motif, and while the TriMV YX-AUG-like motif could be functionally interchanged with variants of both the PV and EMCV YX-AUG motif (Fig. 2), the poor tolerance of the TriMV motif for an extension of spacer sequence and the selection of the first AUG closest to the CU-tract as the preferred start codon (Fig. 2) are more in line with an EMCV-like type II IRES, wherein the TriMV 5′ UTR-mediated translation initiates within the YX-AUG-embedded start codon reached without scanning. Such a model could explain the failure of the native EMCV and PV YX-AUG motifs to support strong TriMV IRES activity in our wheat germ extract and protoplast assays (Fig. 2): either a normally cryptic AUG was recognized as the initiation site and thus prevented translation at the luciferase out-of-frame AUG, or the distance separating the CU-tract from the luciferase initiation site was a limiting factor in the scanning ability of the ribosomal complex (Fig. 2). Nevertheless, it seems clear that both plant and animal viral YX-AUG motifs can exert homologous functions, at least in the context of the elongated TriMV IRES structure.
It is worth noting that the polypyrimidine CU-tract within the picornavirus YX-AUG motif has been reported to be the binding site for the polypyrimidine tract-binding protein (PTB), a crucial interactor for the IRES function (40–42). While it remains possible that the TriMV polypyrimidine CU-tract similarly serves as a binding site for a potential plant PTB homologous protein, based on our functional replacements with diverse 18S rRNA binding sequences (Fig. 2 and 4), our data suggest the possibility for the TriMV YX-AUG motif, wherein it targets and/or positions ribosomal complexes via a tethering mechanism, similar to how Shine-Dalgarno sequences regulate ribosome recruitment to mRNAs in prokaryotic cells (43, 44). While physical base pairing to a 40S complex remains to be demonstrated in most systems, base pairing interactions between mRNAs and various accessible, conserved 18S rRNA stretches have been suggested in several plant viral cap-independent translation enhancers (7), as well as in mammalian and plant cellular mRNAs (36, 45, 46) and mammalian viruses, including poliovirus (47) and hepatitis C virus (48, 49). High-throughput analysis of various 18S rRNA complementary sequences pinpointed the functional region of the 18S rRNA involved in the poliovirus IRES to nt 812 to 1233 (36).
The TriMV YX-AUG motif with its immediately upstream sequences bears two sequence stretches within its CU-tract that shares complementary within that distinct region of the 18S rRNA (nt 1131 to 1140). Our data suggest a redundancy of function for these two complementary stretches because loss-of-function required mutations within either both sites or trans-mediated blockage of both sequences using oligonucleotides (Fig. 3). Similarly, we demonstrate some functional equivalence of bipartite elements from both BRV (Fig. 4) and BVMoV (Fig. 6). While the advantage of redundant signals is not yet clear, particularly since the single 18S rRNA binding site present in the HCRV sequence was sufficient to support TriMV 5′ UTR translation (Fig. 4), one can still speculate that the presence of multiple potential binding sites favors “clustering” of multiple ribosomal subunits on the element at optimal distance to the correct AUG. Similar strategy was proposed for BRV and other related nepoviruses IRES elements (23). It is worth considering that the ability of antisense DNA oligonucleotides to target and inactivate these CU-tracts could inform the development of a potent antiviral strategy based on targeting 18S rRNA binding motifs relevant to both plant and animal viral pathogens.
With the continuous emergence of new potyviruses with distinct 5′ UTR structures (Fig. 4), we speculate that the YX-AUG motif evolved as an advantageous strategy to specify start codons within leader sequences bearing multiple AUGs (Fig. 5). The YX-AUG function is most relevant to IRES-driven translation and is supported by our observations for BVMoV 5′ UTR-driven translation, wherein the BVMoV YX-AUG motif exerted substantial activity but only in the context of the TriMV IRES (Fig. 6). This is the first evidence of a conserved motif among some potyviruses, and it is compelling to think that the BVMoV YX-AUG motif function is either vestigial or only active in the context of specific host determinants or translation strategy. It is worth noting that the inability of the TriMV YX-AUG motif to establish IRES activity of the BVMoV 5′ UTR (Fig. 7C) reveals that, while necessary, the motif alone is not sufficient to drive internal initiation.
In previous work, we have shown that the TriMV IRES requires the translation initiation factor eIF4G or its isoform eIFiso4G, in addition to the DEAD box RNA helicase eIF4A, to drive translation (33). For most picornaviral IRESes, eIF4G and eIF4A may be needed to remodel the IRES near the initiation codon in order to ensure productive binding of the ribosomal complex (9). Although largely dispensable for animal picornaviruses, the viral protein linked to the 5′ end of the genome (VPg) of some potyviruses can favor translation through a direct interaction with the cap-binding factor eIF4E (5). It remains to be determined as to whether the 48.6-kDa TriMV VPg plays such a role in translation. The presence of such bulky protein at the 5′ end of the UTR, which is more than double the size of the typical potyviral VPg (about 20 to 23 kDa), at some stages of the viral life cycle may support the dual function of the TriMV 5′ UTR in translation, with overlapping and distinct sequence requirements, depending on the “free” or “blocked” state of the 5′ end. Indeed, from our previous (11) and current results, we showed that the TriMV 5′ UTR supported cap-independent translation with similar efficiency either in the absence or the presence of the strong hairpin, to presumably block ribosomal scanning and entry from the 5′ end. However, while a 300-nt region of the TriMV 5′ UTR was sufficient to drive translation with a free open 5′ end (11), the full-length UTR was vital for internal initiation to occur in the presence of a 5′-end block (Fig. 1).
In summary, with the similarity of their genome organization with the animal viruses members of the Picornaviridae family, it has been largely assumed that potyviruses share similar mechanism of translation with the animal counterparts. Despite the remarkable diverse translation elements in potyviruses unseen in animal viruses (5), we present here the first evidence of a broad-spectrum translational strategy cross-kingdom.
MATERIALS AND METHODS
Luciferase reporter constructs.
All of the TriMV cDNA clones both in the monocistronic or the dicistronic constructs were generated as described previously (11). The monocistronic TriMV firefly luciferase constructs were assembled in the T3 polymerase-driven plasmid, c-myc-T3LUC(pA) (50). The TriMV control bicistronic constructs were generated from the pDluc plasmid (W. Allen Miller, Iowa State University, Ames, IA). The TriMV mutated sequences were either generated by PCR or synthesized using a gBlocks fragment from IDT-DNA or GenSmart from GenScript. All unrelated sequences, including the Bellflower veinal mottle viral sequence and the human beta-globin sequences, were generated as gBlocks fragment from IDT-DNA or as GenSmart from GenScript and cloned using the In-Fusion cloning kit from Clontech into the TriMV plasmid cut at either HindIII-NcoI sites or HapI-NcoI sites. Our control capped polyadenylated vector construct, designed to mimic a cellular mRNA, was derived from the cmyc-T3Luc (pA) plasmid. After the digestion with HindIII-NcoI, the cut vector was Klenow treated and religated to itself.
Transcription.
All RNAs were transcribed in vitro from linearized plasmids or PCR-amplified products using either the T7 MegaScript kit from Ambion or the T3 or T7 RNA polymerase from Life Technologies. Monocistronic TriMV luciferase constructs and the control luciferase construct with vector sequences were linearized either with SfcI/BfmI to include the poly(A) tail. The dicistronic pDluc-derived constructs were linearized with BamHI to include the poly(A) tail. The renilla luciferase construct (Promega) was linearized with BamHI to include the poly(A) tail.
Reactions were assembled according to the appropriate transcription kit protocol and as previously described (11).
Translation assays and data collection.
The in vitro translation reactions were performed using the wheat germ extract system kit (Promega, Madison, WI) and as previously described (11).
For the translation assays in the presence of the antisense DNA oligonucleotides, luciferase activity was measured for the TriMV 5′ UTR mRNA construct with the strong stem-loop at the immediate 5′ end or for the BVMoV 5′ UTR mRNA construct with an open 5′ end. The translation reaction was set up as previously described but with the addition of 0.6 μl of magnesium acetate to rule out loss of translation due to magnesium chelation by the DNA oligonucleotides. The unmodified DNA oligonucleotides were ordered at IDT-DNA. The sequences were as follow: anti-BoxA, 5′-AAGAGAAGTA-3′; anti-BoxA+jBoxB, 5′-AGGAGAAAAAGAGAAGTA-3′; anti-β-globin, 5′-CAAAAGCTTGCAGGAAGAGATCCATCTAC-3′; anti-BoxA′, 5′-AGAAGTA-3′; anti-BoxB′, 5′-CGAAAGAAA-3′; anti-BoxA′+BoxB′, 5′-CGAAGAAACAGCAAACGACAGAAGTA-3′; and anti-HCRV, 5′-CTGATTCAGCCTTAATCAG-3′.
In vivo translation in oat protoplasts.
Oat protoplasts were prepared from an oat cell suspension culture as described previously (11, 51). A 1-pmol portion of each RNA reporter construct was electroporated into approximately 106 cells. To normalize RNA incorporation into cells, 0.1 pmol of capped polyadenylated renilla luciferase RNA was included in each electroporation. At 5 h postelectroporation, the cells were harvested, lysed in 500 μl of passive lysis buffer (Dual Luciferase kit; Promega), and centrifuged for 10 min at 15,000 × g. Luciferase activity was measured using 100 μl of the supernatant. Then, 50 μl of the luciferase assay reagent was injected into each sample and read for 10 s. Next, 50 μl of Stop&Glo reagent was injected, and the renilla luciferase activity was measured for 6 s. All experiments were performed in triplicate and repeated in at least three independent experiments.
ACKNOWLEDGMENTS
This study was supported by grants from the Hatch Act Formula Fund (MSN169250) and a Wisconsin Alumni Research Foundation Fund (MSN194345) to A.M.R. We declare that we have no conflicts of interest.
REFERENCES
- 1.Jackson RJ, Hellen CU, Pestova TV. 2010. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kneller EL, Rakotondrafara AM, Miller WA. 2006. Cap-independent translation of plant viral RNAs. Virus Res 119:63–75. doi: 10.1016/j.virusres.2005.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martinez-Salas E, Francisco-Velilla R, Fernandez-Chamorro J, Lozano G, Diaz-Toledano R. 2015. Picornavirus IRES elements: RNA structure and host protein interactions. Virus Res 206:62–73. doi: 10.1016/j.virusres.2015.01.012. [DOI] [PubMed] [Google Scholar]
- 4.Reineke LC, Lloyd RE. 2011. Animal virus schemes for translation dominance. Curr Opin Virol 1:363–372. doi: 10.1016/j.coviro.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang J, Roberts R, Rakotondrafara AM. 2015. The role of the 5′ untranslated regions of Potyviridae in translation. Virus Res 206:74–81. doi: 10.1016/j.virusres.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 6.Miras M, Miller WA, Truniger V, Aranda MA. 2017. Noncanonical translation in plant RNA viruses. Front Plant Sci 8:494. doi: 10.3389/fpls.2017.00494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Truniger V, Miras M, Aranda MA. 2017. Structural and functional diversity of plant virus 3′-cap-independent translation enhancers (3′-CITEs). Front Plant Sci 8:2047. doi: 10.3389/fpls.2017.02047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simon AE, Miller WA. 2013. 3′ cap-independent translation enhancers of plant viruses. Annu Rev Microbiol 67:21–42. doi: 10.1146/annurev-micro-092412-155609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Filbin ME, Kieft JS. 2009. Toward a structural understanding of IRES RNA function. Curr Opin Struct Biol 19:267–276. doi: 10.1016/j.sbi.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lozano G, Martinez-Salas E. 2015. Structural insights into viral IRES-dependent translation mechanisms. Curr Opin Virol 12:113–120. doi: 10.1016/j.coviro.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 11.Roberts R, Zhang J, Mayberry LK, Tatineni S, Browning KS, Rakotondrafara AM. 2015. A unique 5′ translation element discovered in triticum mosaic virus. J Virol 89:12427–12440. doi: 10.1128/JVI.02099-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kozak M. 1989. Circumstances and mechanisms of inhibition of translation by secondary structure in eucaryotic mRNAs. Mol Cell Biol 9:5134–5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pelletier J, Kaplan G, Racaniello VR, Sonenberg N. 1988. Cap-independent translation of poliovirus mRNA is conferred by sequence elements within the 5′ noncoding region. Mol Cell Biol 8:1103–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elroy-Stein O, Fuerst TR, Moss B. 1989. Cap-independent translation of mRNA conferred by encephalomyocarditis virus 5′ sequence improves the performance of the vaccinia virus/bacteriophage T7 hybrid expression system. Proc Natl Acad Sci U S A 86:6126–6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duke GM, Hoffman MA, Palmenberg AC. 1992. Sequence and structural elements that contribute to efficient encephalomyocarditis virus RNA translation. J Virol 66:1602–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martínez-Salas E. 2008. The impact of RNA structure on picornavirus IRES activity. Trends Microbiol 16:230–237. doi: 10.1016/j.tim.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wimmer E, Paul AV. 2010. The making of a picornavirus genome, p 33–55. In Ehrenfeld E, Domingo E, Roos R (ed), Picornaviruses. ASM Press, Washington, DC. [Google Scholar]
- 18.Hellen CUT, Pestova TV, Wimmer E. 1994. Effect of mutations downstream of the internal ribosome entry site on initiation of poliovirus protein synthesis. J Virol 68:6312–6322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carrington JC, Freed DD. 1990. Cap-independent enhancement of translation by a plant potyvirus 5′ nontranslated region. J Virol 64:1590–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Basso J, Dallaire P, Charest PJ, Devantier Y, Laliberte JF. 1994. Evidence for an internal ribosome entry site within the 5′ non-translated region of turnip mosaic potyvirus RNA. J Gen Virol 75:3157–3165. doi: 10.1099/0022-1317-75-11-3157. [DOI] [PubMed] [Google Scholar]
- 21.Levis C, Astier-Manifacier S. 1993. The 5′ untranslated region of PVY RNA, even located in an internal position, enables initiation of translation. Virus Genes 7:367–379. doi: 10.1007/BF01703392. [DOI] [PubMed] [Google Scholar]
- 22.Niepel M, Gallie DR. 1999. Identification and characterization of the functional elements within the tobacco etch virus 5′ leader required for cap-independent translation. J Virol 73:9080–9088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karetnikov A, Lehto K. 2007. The RNA2 5′ leader of Blackcurrant reversion virus mediates efficient in vivo translation through an internal ribosomal entry site mechanism. J Gen Virol 88:286–297. doi: 10.1099/vir.0.82307-0. [DOI] [PubMed] [Google Scholar]
- 24.Ivanov PA, Karpova OV, Skulachev MV, Tomashevskaya OL, Rodionova NP, Dorokhov YL, Atabekov JG. 1997. A tobamovirus genome that contains an internal ribosome entry site functional in vitro. Virology 232:32–43. [DOI] [PubMed] [Google Scholar]
- 25.Fernandez-Miragall O, Hernandez C. 2011. An internal ribosome entry site directs translation of the 3’-gene from Pelargonium flower break virus genomic RNA: implications for infectivity. PLoS One 6:e22617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jaag HM, Kawchuk L, Rohde W, Fischer R, Emans N, Prüfer D. 2003. An unusual internal ribosomal entry site of inverted symmetry directs expression of a potato leafroll polerovirus replication-associated protein. Proc Natl Acad Sci U S A 100:8939–8944. doi: 10.1073/pnas.1332697100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khan MA, Yumak H, Gallie DR, Goss DJ. 2008. Effects of poly(A)-binding protein on the interactions of translation initiation factor eIF4F and eIF4F.4B with internal ribosome entry site (IRES) of tobacco etch virus RNA. Biochim Biophys Acta 1779:622–627. doi: 10.1016/j.bbagrm.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 28.Zeenko V, Gallie DR. 2005. Cap-independent translation of tobacco etch virus is conferred by an RNA pseudoknot in the 5′-leader. J Biol Chem 280:26813–26824. doi: 10.1074/jbc.M503576200. [DOI] [PubMed] [Google Scholar]
- 29.Gallie DR, Tanguay RL, Leathers V. 1995. The tobacco etch viral 5′ leader and poly(A) tail are functionally synergistic regulators of translation. Gene 165:233–238. [DOI] [PubMed] [Google Scholar]
- 30.Simon-Buela L, Guo HS, Garcia JA. 1997. Cap-independent leaky scanning as the mechanism of translation initiation of a plant viral genomic RNA. J Gen Virol 78:2691–2699. doi: 10.1099/0022-1317-78-10-2691. [DOI] [PubMed] [Google Scholar]
- 31.Tatineni S, Ziems AD, Wegulo SN, French R. 2009. Triticum mosaic virus: a distinct member of the family Potyviridae with an unusually long leader sequence. Phytopathology 99:943–950. doi: 10.1094/PHYTO-99-8-0943. [DOI] [PubMed] [Google Scholar]
- 32.Seifers DL, Martin TJ, Harvey TL, Fellers JP, Stack JP, Ryba-White M, Haber S, Krokhin O, Spicer V, Lovat N, Yamchuk A, Standing KG. 2008. Triticum mosaic virus: a new virus isolated from wheat in Kansas. Plant Dis 92:808–817. doi: 10.1094/PDIS-92-5-0808. [DOI] [PubMed] [Google Scholar]
- 33.Roberts R, Mayberry LK, Browning KS, Rakotondrafara AM. 2017. The triticum mosaic virus 5′ leader binds to both eIF4G and eIFiso4G for translation. PLoS One 12:e0169602. doi: 10.1371/journal.pone.0169602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seo JK, Kwak HR, Kim MK, Kim JS, Choi HS. 2017. The complete genome sequence of a novel virus, Bellflower veinal mottle virus, suggests the existence of a new genus within the family Potyviridae. Arch Virol 162:2457–2461. doi: 10.1007/s00705-017-3374-5. [DOI] [PubMed] [Google Scholar]
- 35.Kaminski A, Belsham GJ, Jackson RJ. 1994. Translation of encephalomyocarditis virus RNA: parameters influencing the selection of the internal initiation site. EMBO J 13:1673–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weingarten-Gabbay S, Elias-Kirma S, Nir R, Gritsenko AA, Stern-Ginossar N, Yakhini Z, Weinberger A, Segal E. 2016. Comparative genetics: systematic discovery of cap-independent translation sequences in human and viral genomes. Science 351:aad4939-1. doi: 10.1126/science.aad4939. [DOI] [PubMed] [Google Scholar]
- 37.Wang Z, Kraft JJ, Hui AY, Miller WA. 2010. Structural plasticity of Barley yellow dwarf virus-like cap-independent translation elements in four genera of plant viral RNAs. Virology 402:177–186. doi: 10.1016/j.virol.2010.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koh DC, Wong SM, Liu DX. 2003. Synergism of the 3′ untranslated region and an internal ribosome entry site differentially enhances the translation of a plant virus coat protein. J Biol Chem 278:20565–20573. doi: 10.1074/jbc.M210212200. [DOI] [PubMed] [Google Scholar]
- 39.Seo JK, Kwak HR, Lee YJ, Kim J, Kim MK, Kim CS, Choi HS. 2015. Complete genome sequence of bellflower vein chlorosis virus, a novel putative member of the genus Waikavirus. Arch Virol 160:3139–3142. doi: 10.1007/s00705-015-2606-9. [DOI] [PubMed] [Google Scholar]
- 40.Kafasla P, Morgner N, Robinson CV, Jackson RJ. 2010. Polypyrimidine tract-binding protein stimulates the poliovirus IRES by modulating eIF4G binding. EMBO J 29:3710–3722. doi: 10.1038/emboj.2010.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jang SK, Wimmer E. 1990. Cap-independent translation of encephalomyocarditis virus RNA: structural elements of the internal ribosomal entry site and involvement of a cellular 57-kD RNA-binding protein. Genes Dev 4:1560–1572. [DOI] [PubMed] [Google Scholar]
- 42.Kaminski A, Jackson RJ. 1998. The polypyrimidine tract binding protein (PTB) requirement for internal initiation of translation of cardiovirus RNAs is conditional rather than absolute. RNA 4:626–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shine J, Dalgarno L. 1974. The 3’-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A 71:1342–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kozak M. 1999. Initiation of translation in prokaryotes and eukaryotes. Gene 234:187–208. [DOI] [PubMed] [Google Scholar]
- 45.Scharff LB, Ehrnthaler M, Janowski M, Childs LH, Hasse C, Gremmels J, Ruf S, Zoschke R, Bock R. 2017. Shine-Dalgarno sequences play an essential role in the translation of plastid mRNAs in tobacco. Plant Cell 29:3085–3101. doi: 10.1105/tpc.17.00524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martin F, Menetret JF, Simonetti A, Myasnikov AG, Vicens Q, Prongidi-Fix L, Natchiar SK, Klaholz BP, Eriani G. 2016. Ribosomal 18S rRNA base pairs with mRNA during eukaryotic translation initiation. Nat Commun 7:12622. doi: 10.1038/ncomms12622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pilipenko EV, Gmyl AP, Maslova SV, Svitkin YV, Sinyakov AN, Agol VI. 1992. Prokaryotic-like cis elements in the cap-independent internal initiation of translation on picornavirus RNA. Cell 68:119–131. [DOI] [PubMed] [Google Scholar]
- 48.Malygin AA, Kossinova OA, Shatsky IN, Karpova GG. 2013. HCV IRES interacts with the 18S rRNA to activate the 40S ribosome for subsequent steps of translation initiation. Nucleic Acids Res 41:8706–8714. doi: 10.1093/nar/gkt632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matsuda D, Mauro VP. 2014. Base pairing between hepatitis C virus RNA and 18S rRNA is required for IRES-dependent translation initiation in vivo. Proc Natl Acad Sci U S A 111:15385–15389. doi: 10.1073/pnas.1413472111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thoma C, Bergamini G, Galy B, Hundsdoerfer P, Hentze MW. 2004. Enhancement of IRES-mediated translation of the c-myc and BiP mRNAs by the poly(A) tail is independent of intact eIF4G and PABP. Mol Cell 15:925–935. doi: 10.1016/j.molcel.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 51.Roberts R, Zhang J, Mihelich N, Savino D, Rakotondrafara AM. 2017. Manipulation of oat protoplasts for transient expression assays. Methods Mol Biol 1536:55–70. doi: 10.1007/978-1-4939-6682-0_5. [DOI] [PubMed] [Google Scholar]