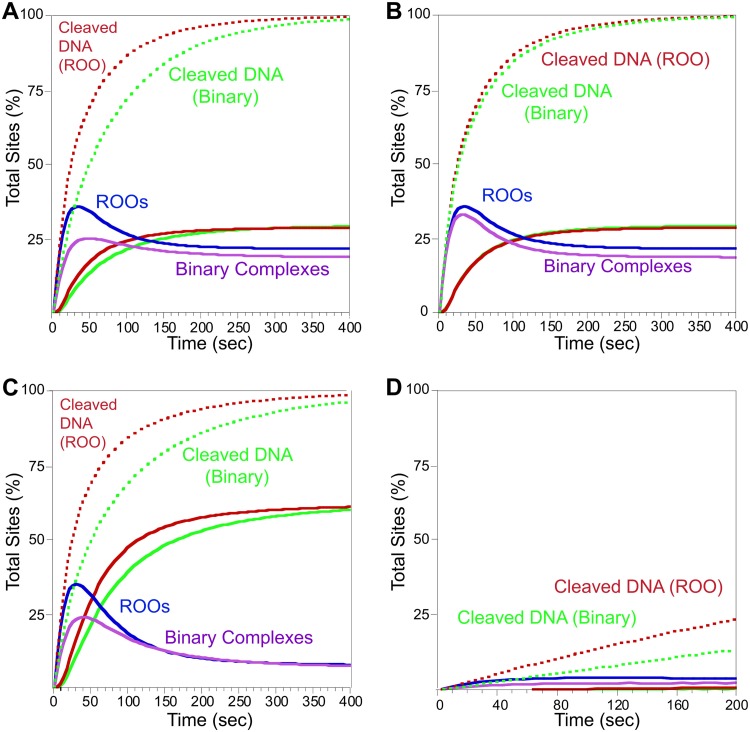
FIG 5.
Model of phage DNA cleavage and comparison of ROO and binary mechanisms. Cleaved DNA bound to SgrAI or the binary enzyme is shown in dotted red (ROO mechanism) or green (binary mechanism). Cleaved DNA released from SgrAI or the binary enzyme is shown in solid lines (red, ROO mechanism; green, Binary). Concentrations of ROO filaments (with cleaved and uncleaved DNA) are blue, and Binary complexes are purple. See Tables S3 and S4 for model details and rate constants. (A) Simulation of DNA sites in phage DNA. Starting concentration was 80 nM DNA, the estimated local concentration of SgrAI bound to two primary sites present on the same DNA molecule. (B) As for panel A, but the Binary non-ROO mechanism is now set to allow for two ways to form assemblies. (C) As for panel A, but with rebinding of cleaved DNA set to a lower off-rate constant to mimic the lower concentrations of DNA free in the cell (see the text for details). (D) As for panel A, but with 3 nM DNA to mimic reactions between separate molecules in the cell (i.e., host genome and phage DNA).