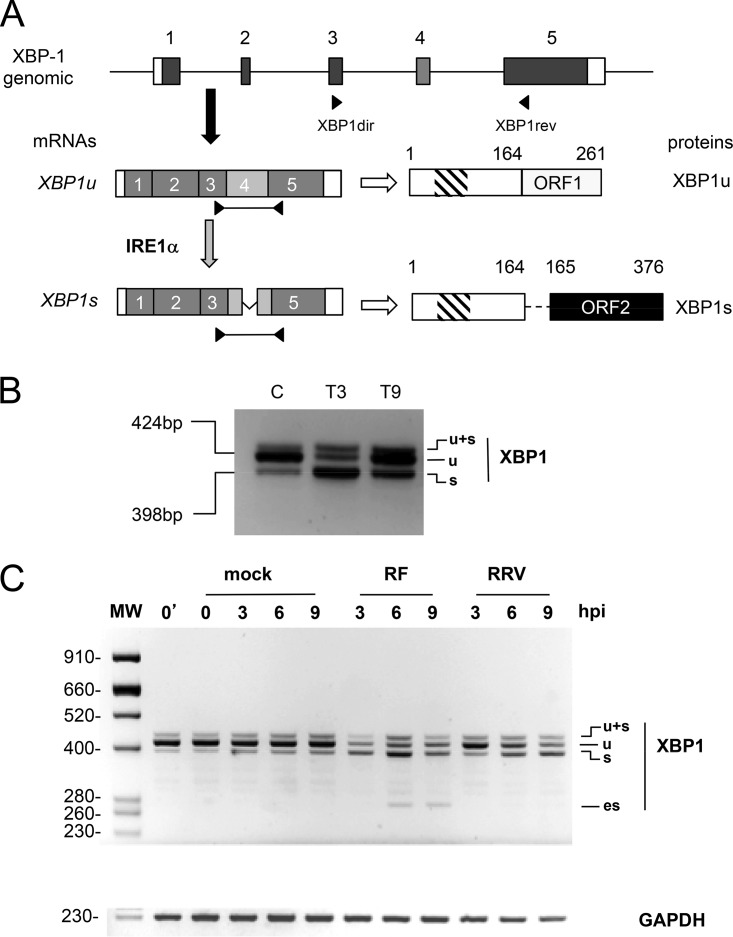
FIG 1.
Rotavirus infection induces cytoplasmic splicing of XBP1. (A) Schematic structures of the XBP1u and XBP1s mRNAs. The general organization of the XBP1 gene is indicated with exons as gray boxes, noncoding regions are indicated as white boxes, and introns are indicated as lines (not to scale). The mRNA encoding XBP1u is produced by canonical nuclear splicing and then exported to the cytoplasm (black arrow) to be translated (white arrow). The XBP1u protein translated from XBP1u mRNA is 261 amino acids long and contains a DNA-binding domain (hatched box). The mRNA encoding XBP1s is produced by an unconventional cytoplasmic splicing (gray arrow) of XBP1u mRNA at exon 4 (light gray) catalyzed by the IRE1α endoribonuclease. The XBP1s protein is 376 amino acids long and contains DNA-binding and transactivating (black box) domains. (B) Detection of XBP1u splicing by IRE1α. The positions of the primers XBP1dir and XBP1rev on the XBP1 gene and mRNAs are indicated. The RT-PCR products (424 and 398 bp) obtained using these primers on RNA purified from unstressed MA104 cells (lane C) or MA104 cells treated with thapsigargin (400 nM) for 3 and 9 h (T3, T9) are illustrated. (C) The DNA products obtained by RT-PCR from RNA extracted from mock-infected cells or from cells infected (MOI of 10) with rotavirus RF or RRV for the indicated time (in hours) were analyzed by agarose gel electrophoresis. The top panel shows the XBP1 RT-PCR products, and the bottom panel shows the GAPDH RT-PCR products used as a loading control. The 0′ lane corresponds to untreated cells, and the 0 lane corresponds to mock-infected cells. The sizes of the molecular weight markers (MW) are indicated in base pairs on the left side.