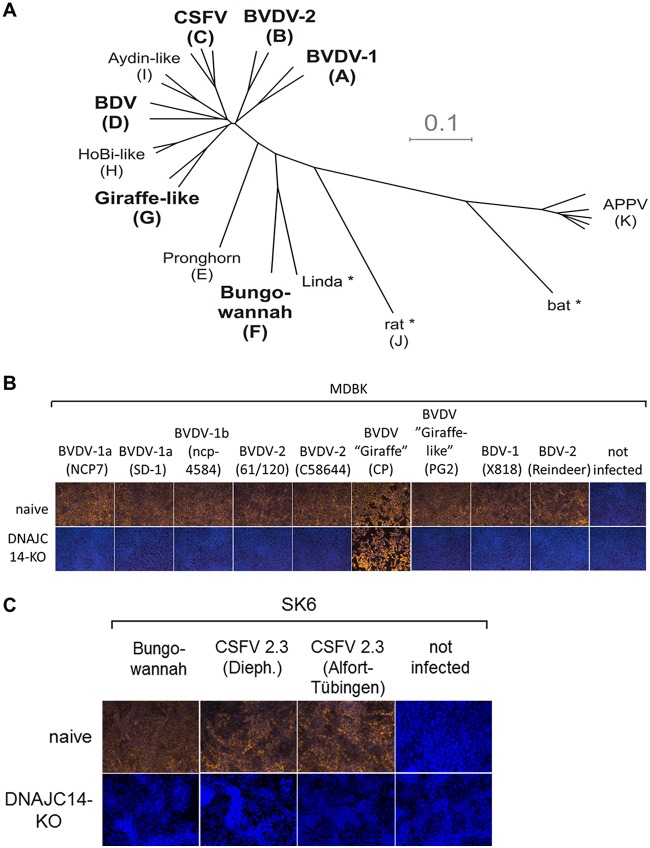
FIG 4.
Infection of DNAJC14-KO cells demonstrates that replication of noncp pestiviruses depends on DNAJC14. (A) Phylogenetic tree (modified from reference 109) representing the four established pestivirus species BVDV-1, BVDV-2, BDV, and CSFV, together with the additional tentative pestivirus species recently proposed by the Flaviviridae Study Group of the International Committee on Taxonomy of Viruses (71). Letters in brackets represent the pestivirus species Pestivirus A to K according to the recent proposal for a revised pestivirus nomenclature (71). Pestivirus species used for the experiments in panel B are BVDV-1, BVDV-2, BDV, CSFV, giraffe, and Bungowannah (highlighted in boldface letters). Asterisks indicate pestiviruses for which no isolates exist or are not available. (B) Naïve MDBK (upper) and MDBK DNAJC14-KO clone 2 cells (lower) were infected with noncp pestivirus isolates BVDV-1b NCP7, BVDV-1a SD-1, BVDV-1b ncp-4584, BVDV-2 61/120, BVDV-2 C58644, and PG-2, the latter of which belongs to the pestivirus species giraffe (pestivirus G), BDV-1 X818, BDV-2 reindeer, and, as a control, cp strain giraffe (pestivirus G). (C) Naïve SK6 (upper) and SK6 DNAJC14-KO clone 9 (lower) cells were infected with noncp pestivirus CSFV strains Alfort-Tübingen and Diepholz (Dieph.) and noncp pestivirus strain Bungowannah. All infections were performed in parallel in the respective DNAJC14-KO cell lines with the indicated pestivirus isolates at an MOI of 1. Viral genome replication was monitored by the anti-NS3-based IF assay at 72 hpi (depicted as yellow fluorescence). Cell nuclei were counterstained with DAPI. All infections were performed in three independent experiments, and a representative data set is shown.