FIG 3.
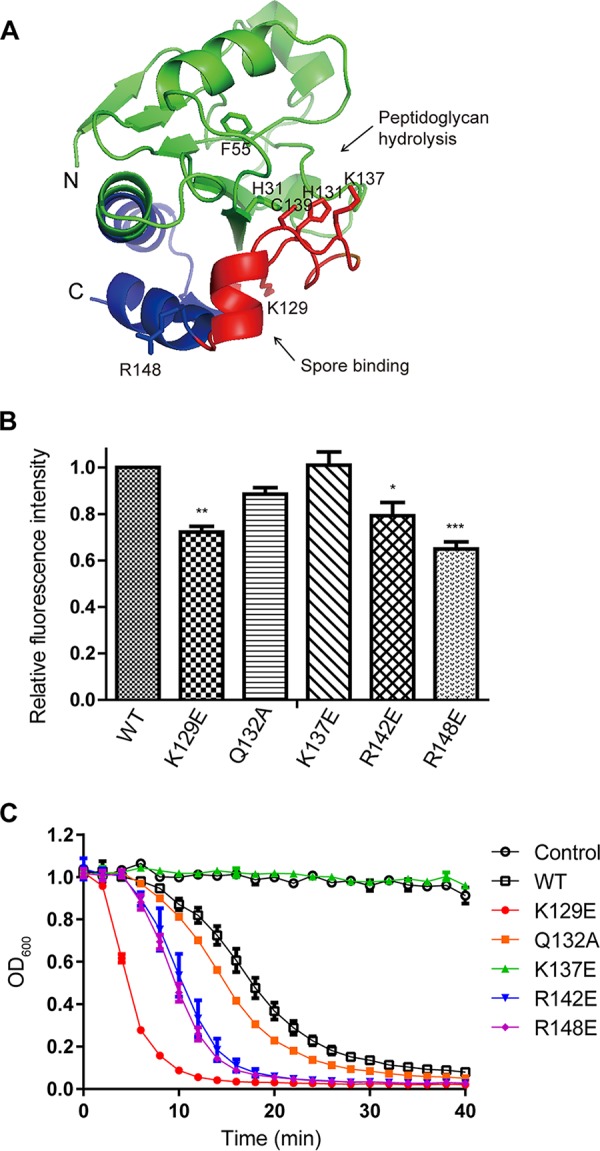
The effects of point mutations within SBD on the activity of LysPBC2. (A) Structural model of the catalytic domain of LysPBC2 (residues 1 to 158). The SBD region is represented in blue with the highly conserved core region (residues 127 to 147) in red. The N and C termini are marked N and C, respectively. Catalytic residues (H31, F55, H131, K137, and C139) and key residues for spore binding (K129 and R148) are shown as sticks. (B) Effects of point mutations within the SBD on spore binding. *, P < 0.05; **, P < 0.01; ***, P < 0.001 versus the control (WT), determined by one-way ANOVA with Tukey’s post hoc test. (C) Effects of point mutations within the SBD on the lytic activity of LysPBC2 against vegetative cells.
