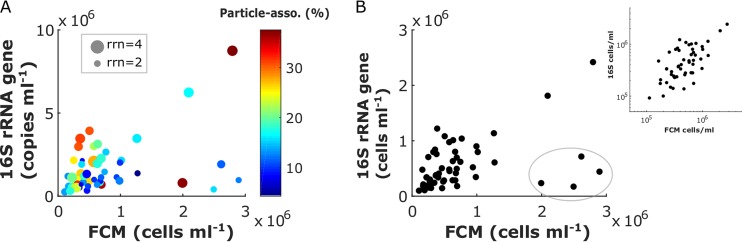
FIG 4.
Comparison of total bacterial abundances estimated by FCM and 16S rRNA gene QMP. (A) The size of each data point represents the rrn effect, calculated as the averaged rrn of the top 20 classified OTUs in each sample. The color coding represents the particle association effect, calculated as the cumulative cell percentage (after rrn correction) of the top 10 most abundant particle-associated OTUs, identified as the >3-µm fraction bacteria reported by Delmont et al. (41). An exhaustive survey of the particle-associated OTUs is not feasible considering that significant portions of the prokaryotic OTUs are of unknown physiology or are even unclassified in the current rRNA database (SILVA 128). (B) FCM versus rrn and particle association effect-corrected 16S rRNA QMP in cells milliliter−1. The four points inside the gray circle are likely outliers. After excluding these four points, data points are plotted on both linear and log scales.