Figure 2. Bifidobacterium strains in DIABIMMUNE children.
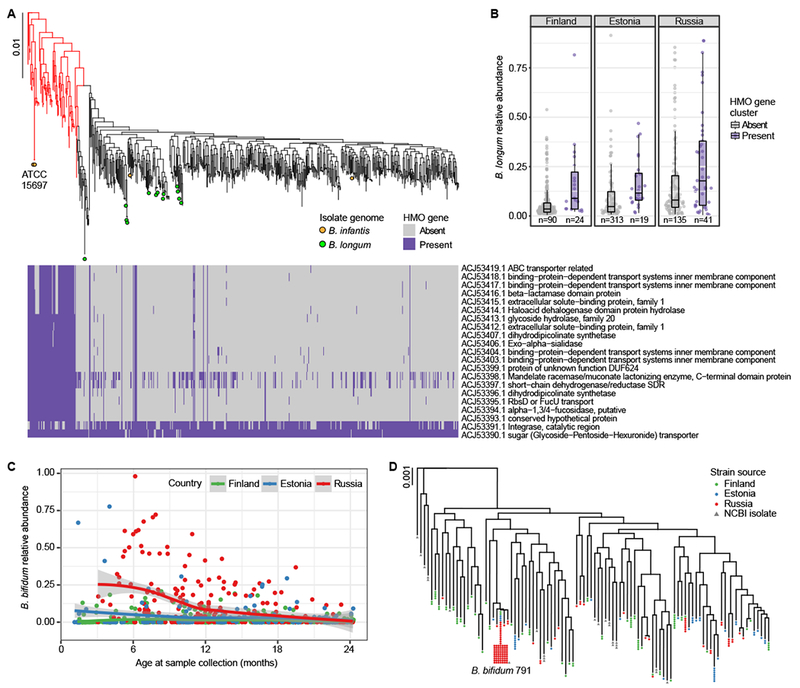
A Phylogenetic tree of B. longum strains in DIABIMMUNE stool samples and 18 NCBI B. longum isolate genomes based on SNP haplotypes. Highlighted B. infantis strains (red) include two reference sequences (ATCC 15697). The heatmap illustrates strain-specific carriage of 21 genes in the B. infantis HMO gene cluster responsible for intracellular HMO degradation, evaluated using the metagenomic data. B B. longum relative abundance stratified by country and B. longum strain; B. infantis (highlighted red in panel A) has, on average, higher relative abundance compared to other B. longum strains (mixed effects logistic regression p=0.00049). The box shows the interquartile range (IQR), the vertical line shows the median and the whiskers show the range of the data (up to 1.5 times IQR). Number of samples (n) are indicated below each box and include samples from subjects with no breastfeeding information. C Relative abundance of B. bifidum longitudinally stratified by the countries up to 24 months of age (n = 864). Russians have more B. bifidum, especially during the first year of life. The curves show locally weighted scatterplot smoothing (LOESS) fits for the relative abundances, and shaded areas show 95% confidence interval for each fit, as implemented in geom_smooth function in ggplot2 R package. D Phylogenetic tree of B. bifidum strains in the DIABIMMUNE stool samples based on SNP haplotypes. Strains with >99.5% sequence similarity have been collapsed into a single tip. A known strain, B. bifidum 791, was found in 79 stool samples. The scale bars on phylogenetic trees denote difference in sequence similarity of SNP haplotypes.
