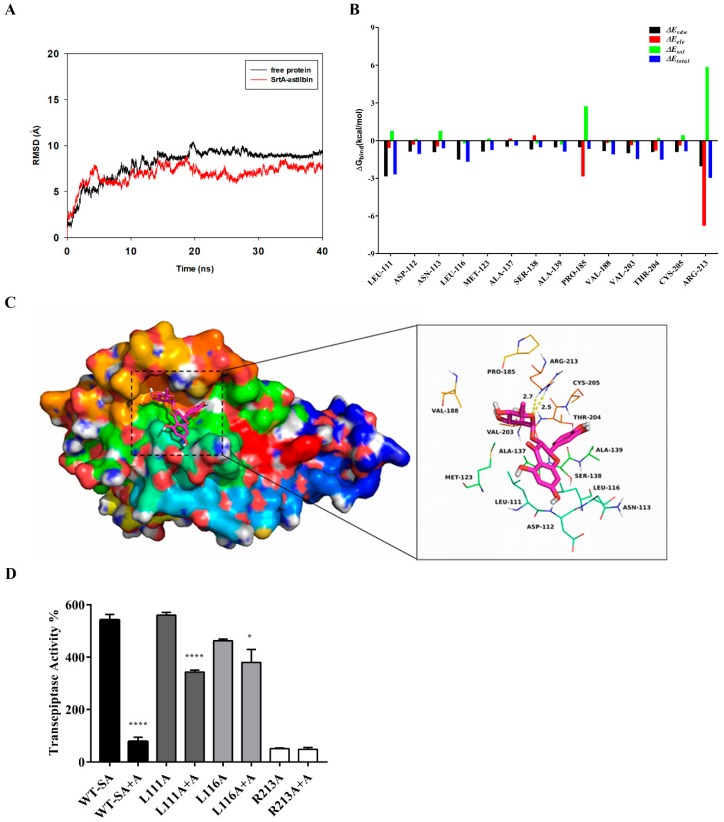
Figure 4.
The results of the molecular docking (MD) simulation of the SrtA-astilbin complex. (A) The root-mean-square deviations (RMSD) exposed by the backbone atoms of the protein during the MD simulation of the SrtA-astilbin complex. (B) Decomposition of the binding free energy on a per residue basis in the SrtA-astilbin complex. (C) The predicted interaction mode of astilbin with the amino acid residues of the catalytic center of SrtA. (D) The inhibition effect of astilbin (64 μg/mL) (A represents astilbin) on the activities of SrtA and its mutants L111A-SrtA, L116A-SrtA, and R213A-SrtA. Significant differences between groups were accepted at * p < 0.05 and **** p < 0.0001.