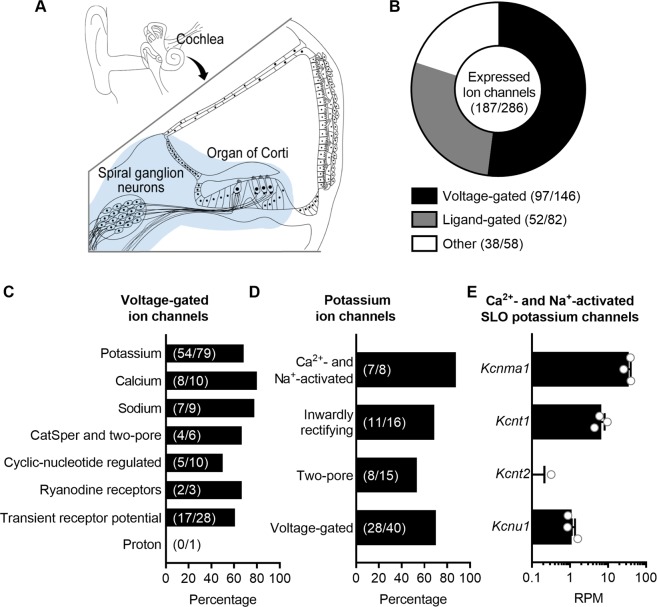
Figure 1.
RNAseq identifies transcripts encoding the SLO channels KCa1.1, KNa1.1, KNa1.2 but not KCa5.1 in sensorineural structures of the mouse inner ear. (A) RNAseq was used to obtain whole transcriptomes from the sensorineural structures of the cochlea, including organs of Corti and spiral ganglion neurons (blue highlighted area), from post-hearing 6-week-old mice. (B) Following the classification of the IUPHAR/BPS Guide to Pharmacology, transcripts corresponding to a total of 65% of known ion channels (187/286 genes), subdivided into voltage-gated, ligand-gated and other ion channels, are expressed in the sensorineural structures. (C) The majority of transcripts encoding voltage-gated ion channels encode potassium channels, with transcripts corresponding to a total of 68% of known potassium channels (54/79 genes) expressed. (D) Of the potassium channels, 88% of the Ca2+- and Na+-activated potassium channels (7/8 genes) are expressed. (E) Three of the four members of the subset of Ca2+- and Na+-activated potassium channels encoding the SLO family of ion channels are expressed. For this group of ion channels, Kcnma1, which encodes KCa1.1/SLO1/BK, is most abundantly expressed. Kcnt1, which encodes KNa1.1/SLO2.2/Slack, is expressed at intermediate levels. Kcnt2, which encodes KNa1.2/SLO2.1/Slick, is absent from two of the three replicates and expressed at <1 RPM in one replicate. Kcnu1, which encodes KCa5.1/SLO3, is expressed at low levels. Data are plotted to show individual replicates (animals) and mean ± SEM. Values (mean ± SEM) are provided in the Results.