Abstract
Microbial fermentation for L-methionine (L-Met) production based on natural renewable resources is attractive and challenging. In this work, the effects of medium composition and fermentation conditions were investigated to improve L-Met production by genetically engineered Escherichia coli MET-3. Statistical optimization techniques including Plackett–Burman (PB) design and Box–Behnken design (BBD) were adopted first to optimize the culture medium. Results of PB-designed experiments indicated that the culture medium components including glucose, yeast extract, KH2PO4, and MgSO4.7H2O had significant effects on L-Met biosynthesis. With their best-predicted concentration established by BBD (glucose 37.43 g/L, yeast extract 0.95 g/L, KH2PO4 1.82 g/L, and MgSO4.7H2O 4.51 g/L), L-Met titer was increased to 3.04 g/L from less than 2.0 g/L. For further enhancement of L-Met biosynthesis, the fermentation conditions of batch cultivation carried out in a 5-L fermentor were optimized, and the optimum results were obtained at an agitation rate of 300 rpm, medium pH of 7.0, and induction temperature of 28 °C. Based on the optimization parameters, fed-batch fermentation with the modified medium was conducted. As a result, great improvement of L-Met titer (12.80 g/L) and yield (0.13 mol/mol) were achieved, with an increase of 38.53% and 30.0% compared with those of the basal medium, respectively. Furthermore, higher L-Met productivity of 0.261 g/L/h was obtained, representing 2.13-fold higher in comparison to the original medium. The results may provide a helpful reference for further study on strain improvement and fermentation control.
Electronic supplementary material
The online version of this article (10.1007/s13205-019-1609-8) contains supplementary material, which is available to authorized users.
Keywords: L-Methionine, Optimization, Escherichia coli, Plackett–Burman, Box–Behnken design
Introduction
Methionine (Met) is a sulfur-containing essential amino acid required in the diet of humans, other mammals, and avian species for normal growth and functions of the body (Chen et al. 2014; Jankowski et al. 2014; Kumar et al. 2003). Met plays indispensable roles in multiple cellular processes and activities, including synthesis, stability, structure, regulation of catalytic function, and posttranslational modification of many proteins (Maresi et al. 2015). In addition to being an initiating amino acid in eukaryotic polypeptide chain synthesis and a precursor for all other sulfur-containing amino acids and their derivatives, it serves as the major methyl group donor in vivo, including the methyl groups for DNA and RNA intermediates (Gomes and Kumar 2005; Jankowski et al. 2014). As an important amino acid, Met is widely used in feed, pharmaceutical and food industries (Figge et al. 2016; Li et al. 2017).
In present years, the demand of Met has gradually increased due to the rapid growth of feed additive market driven by the globally increasing consumption of meat and milk products as a source of protein and other nutrients (Willke 2014). Met can be produced in chemical routes or biological routes. To date, almost all the commercial production of Met relies on chemical synthesis. However, chemical routes for Met synthesis not only produce a racemic mixture of D- and L-forms but also result in serious environmental pollution (Gomes and Kumar 2005; Li et al. 2017). Therefore, a low-cost, environmentally friendly production of pure L-Met with microorganisms based on natural renewable resources is becoming more attractive (Willke 2014).
The L-Met biosynthetic pathway and regulatory mechanisms have been extensively studied and well known in many microorganisms, particularly in Escherichia coli (Li et al. 2017; Mondal et al. 1996; Usuda and Kurahashi 2005; Weissbach and Brot 1991) and Corynebacterium glutamicum (Kase and Nakayama 1975; Mampel et al. 2005; Park et al. 2007). Over the past few decades, large amounts of work were devoted on screening and mutagenesis of microorganism strains for L-Met over-production and obtained great academic results and achievements. Nevertheless, the microbial fermentation-based L-Met production is still challenging and an industrial application has not yet been developed, despite its high commercial interest (Teleki et al. 2017). This is largely because, L-Met biosynthetic pathway is highly branched with divergent and convergent sections and the metabolic regulation is strict and complicated (Kase and Nakayama 1975; Li et al. 2017). Thus, more targeted modifications of its metabolic pathway and feed-back regulation with rational metabolic engineering strategies are necessary to construct new L-Met over-producers. In our previous work, a series of L-Met producing strains (E. coli W3110 derivatives) were constructed based on the comprehensive analysis of feedback inhibition, synthetic bottleneck, and cell growth repression in L-Met biosynthesis, and L-Met production were improved with different extents (Huang et al. 2017, 2018a, b).
Apart from the strain improvement, optimization of nutritional components and culture conditions are essential to enhance the output of the target product during the fermentation process. As to optimization techniques, a number of modeling and optimization methodologies are available. Among those, statistical experimental designs Plackett–Burman (PB) design and Box–Behnken design (BBD) are effective, powerful approaches for rapid screening of key factors from a multi-variable system and prove more advantageous over the conventional one-factor-at-a-time (OFAT) strategy (Dayana Priyadharshini and Bakthavatsalam 2016). The PB design is a preliminary optimization technique used to unbiasedly estimate the effect of all variables through a few experiments (Ma et al. 2016). BBD is an extensively used approach of response surface methodology (RSM), allowing for reduced data points compared with a Central Composite design (CCD) (Sibanda and Pretorius 2012). RSM is a combination of mathematical and statistical techniques used to analyze the effects and interactions of the variables, which has been successfully applied to optimize the biochemical processes (Dayana Priyadharshini and Bakthavatsalam 2016; Zou et al. 2015). In contrast to conventional methods, RSM can evaluate the interaction among several variables to obtain one or more optimal response by mathematically modeling, even when little is known about the process (Asadi and Zlouei 2017; Hou et al. 2019; Shahbazmohammad and Omidinia 2017).
The present work was performed to design suitable fermentation medium and optimize the cultivation conditions for achieving high L-Met production by the genetically engineered E. coli strain MET-3 constructed in our previous work (Huang et al. 2018a). In medium optimization process, different nutritional components such as carbon, nitrogen, and sulphur sources that potentially influence L-Met biosynthesis were investigated by means of the PB design to initially screen the key factors for further optimization using RSM based on BBD. Further attempt was made to optimize the fermentation conditions including temperature, pH, and agitation in a 5-L fermentor. Based on the optimization results, fed-batch fermentation was conducted to develop an efficient fermentation process for enhanced L-Met production.
Materials and methods
Bacterial strain and plasmid
A genetically engineered E. coli MET-3 harboring a recombinant plasmid pTrc99A-metA**-yjeH-serAm, previously constructed and preserved in our laboratory was used as L-Met producing strain in this study. In plasmid pTrc99A-metA**-yjeH-serAm, the genes metA (encoding homoserine O-succinyltransferase with reduced feedback sensitivity to S-adenosylmethionine (SAM) and L-Met), yjeH (encoding L-Met efflux transporter), and serAm (encoding mutant phosphoglycerate dehydrogenase) were cloned under the control of the trc promoter in plasmid pTrc99A for enhanced expression of key enzymes involved in L-Met biosynthetic pathway (Huang et al. 2017, 2018a). Ampicillin (Amp) resistance was employed as a selection marker for plasmid-containing cells.
Chemicals
L-Met standard, L-lysine, vitamin B1 (Vb1), vitamin B12 (Vb12), 3,5-dinitrosalicylic acid (DNS), and 2,4-dinitrofluorobenzene (DNFB) were purchased from J&K Scientific Ltd. (Beijing, China). Amp, isopropyl-β-d-thio-galactopyranoside (IPTG), and ethylenediaminetetraacetic acid (EDTA) were provided by Sangon Biotech Co., Ltd. (Shanghai, China). Sodium acetate was provided by Wenzhou Jixiang Chemical Co., Ltd. (Wenzhou, China). Methanol was HPLC grade and purchased from Tedia Company, Inc. (Fairfield, OH, USA). All other chemicals and reagents otherwise demonstrated were of analytical grade and purchased from commercial sources.
Media
The seed medium was Luria–Bertani (LB) medium (tryptone 10 g/L, yeast extract 5 g/L, and NaCl 10 g/L) supplemented with 100 µg/mL Amp.
The basal fermentation medium, slightly modified according to Dischert et al. (2016) was consisted of: 20 g/L glucose, 3.3 g/L (NH4)2SO4, 2.0 g/L yeast extract, 2.5 g/L KH2PO4, 3.95 g/L Na2S2O3, 1.7 g/L citric acid, 1.0 g/L MgSO4.7H2O, 2 mL/L salt solution, 10 g/L CaCO3, 1.0 mg/L Vb12, 1.0 mg/L Vb1, 50 mg/L L-Lys, 100 µg/mL Amp, and 0.1 mM IPTG.
Salt solution contained (g/L): ZnSO4·H2O 8.5, MnCl2·4H2O 7.5, CuCl2·2H2O 0.75, CoCl2·6H2O 1.25, EDTA 4.0, H3BO3 3.0, NaMoO4·2H2O 2.5, FeCl3·6H2O 60, and CaCl2·2H2O 40.
Feeding medium contained: glucose 500 g/L, MgSO4·7H2O 5 g/L, salt solution 2 mL/L, (NH4)2SO4 33 g/L, Na2S2O3 39.5 g/L, VB12 1 mg/L, VB1 1 mg/L, L-Lys 2 g/L, Amp 100 mg/L, and IPTG 1 mM.
Culture conditions
For seed preparation, the recombinant E. coli MET-3 cells were routinely cultured in LB medium containing 100 µg/mL Amp at 37 °C and 180 rpm for 12–16 h, in an orbital incubator shaker ZWYR-D2402 (Zhicheng Analysis Instrument Manufacturing Co., Ltd., Shanghai, China).
The shake flask fermentations for L-Met production with recombinant E. coli MET-3 were carried out in 500-mL Erlenmeyer flasks containing 30 mL basal fermentation medium. The medium was inoculated with pre-inoculums at 5% (v/v) level and cultured at 37 °C and 180 rpm. When OD600 of the cultures reached 0.6–0.8, 0.1 mM IPTG was added to induce enzymes expression (Liu et al. 2017b). The fermentation lasted for 24–48 h at 28 °C and 180 rpm.
The fermentor fermentation was conducted in a 5-L jar fermentor (Winpact Parallel Fermentation System FS-05, Major Science, Taiwan R.O.C.) with a working volume of 3 L for batch fermentation and 2 L for fed-batch fermentation, respectively. The cultivation was initiated by inoculating 5% (v/v) of seed broth. The pH was automatically controlled with 20% (v/v) NH3.H2O and 1 M H3PO4. Cells were grown at 37 °C until OD600 reached about 2.0, and then induced by addition of 0.1 mM IPTG. During the entire fermentation process, the dissolved oxygen (DO) level was maintained above 20% by controlling the agitation rate at 200–900 rpm and aeration of 1.5 vvm (air volume/culture volume/min). For fed-batch fermentation, when initial glucose was depleted, feeding medium was intermittently fed into the fermentor using a computer-controlled pump to keep glucose concentration at approximately 1–10 g/L with pulse feeding mode. The culture was sampled at regular intervals for assay.
Plackett–Burman design and data analysis
The PB design is an efficient screening method to identify the critical factors among a large number of factors that influences a process in a minimal number of runs with good degree of accuracy (Plackett and Burman 1946; Rahman et al. 2010). To define the effects of the main medium composition on L-Met production by E. coli MET-3, the independent effect of eight variables, including glucose, (NH4)2SO4, yeast extract, Na2S2O3, salt solution, KH2PO4, MgSO4.7H2O, and citric acid on the response (L-Met titer) were first tested by a PB screening design. 12 experimental runs and 6 central point replications were carried out to estimate the experimental error and screen significant components affecting L-Met biosynthesis. All experiments were performed in triplicate, and the mean and standard deviations were determined. The experimental data were statistically analyzed with the software package Minitab Release 16 (Minitab Inc., State College, PA, USA). The significant variables were considered based on the F test and the p value at a significance level of 5% (p < 0.05).
Box–Behnken design and statistical analysis
The BBD of RSM was used to optimize the four most significant variables (glucose, yeast extract, KH2PO4, and MgSO4.7H2O), identified by PB experiment. The four independent variables were investigated at three different levels (− 1, 0, and 1) in a set of 24 experiment runs and 5 central point replications. All experiments were carried out in triplicates and the mean L-Met titer (g/L) was taken as the response (Y). The experiment results were analyzed by Design Expert software (Version 8.0.5, Stat-Ease Inc., Minneapolis, USA) and the quadratic regression model was established relating the response to the variables, as described by Eq. (1):
| 1 |
where Y is the predicted L-Met titer (g/L), and for the independent variables of the coded value; is intercept; is the linear coefficient; is square coefficient; (i ≠ j) is interaction coefficient (Ye et al. 2016).
Analytical methods
Biomass was measured based on the optical density at 600 nm (OD600) with an Eppendorf BioPhotometer (Eppendorf AG, Hamburg, Germany) and converted to dry cell weight (DCW) using a calibration Eq. 1 OD600 = 0.31 g/L DCW. The residual glucose was detected using the DNS assay (Lee et al. 2017; Miller 1959). The concentration of L-Met in culture broth were determined by Hitachi Primaide HPLC System (Hitachi, Tokyo, Japan) equipped with a 1410 UV detector (Primaide, Hitachi, Tokyo, Japan) (set at 365 nm) and a reversed phase column Unitary C-18 (4.6*150 mm, 5 µm) (Acchrom, Wenling, China) after pre-column derivatization with DNFB (Zhang et al. 2012). The mobile phase was 25 mM sodium acetate solution (pH 7.8 adjusted by HAc) and methanol (50:50, v/v) at the flow rate of 1.0 mL/min. The column temperature was 40 °C.
Results and discussion
Screening significant medium compositions for L-Met biosynthesis
The medium composition generally displays an important role in microbial fermentation processes, and is one of the main factors affecting the yield of target metabolites (Bicas et al. 2008; Labbeiki et al. 2014). Based on our preliminary results of single-factor tests on the medium component, glucose, (NH4)2SO4, yeast extract, Na2S2O3, salt solution, KH2PO4, MgSO4.7H2O, and citric acid were considered as the main medium component in this study (Suppl. Table S1). To screen the significant factors influencing L-Met biosynthesis by recombinant E. coli MET-3, the data of PB-designed experiments with eight variables were statistically analyzed; the estimated coefficients and significant levels for the variables in L-Met biosynthesis was presented in Table 1. The R2 value was 0.915, indicating that 91.5% of the variability in the response could be explained by the model. The F value and p value were selected for analysis of variance (ANOVA) of the model. According to the statistical analysis results, four variables including glucose, yeast extract, KH2PO4, and MgSO4.7H2O were significant model terms at the 98% level with p values of less than 0.0200 (Table 1). Positive value in the model for a response represents an effect that favors and negative value indicates an inverse relationship between response and a factor (Rahman et al. 2010).
Table 1.
Significance analysis of eight variables for L-Met biosynthesis from the PB design results
| Variable | Effect | Coef | SE Coef | T | F | P value Prob. > F |
|---|---|---|---|---|---|---|
| Model | 1.0549 | 0.04111 | 25.66 | 9.11 | 0.000 | |
| x 1 (glucose) | 0.4242 | 0.2121 | 0.04111 | 5.16 | 26.62 | 0.001*** |
| x 2 ((NH4)2SO4) | − 0.1260 | − 0.0630 | 0.04111 | − 1.53 | 2.35 | 0.164 |
| x 3 (yeast extract) | − 0.2381 | − 0.1190 | 0.04111 | − 2.90 | 8.39 | 0.020* |
| x 4 (Na2S2O3) | − 0.1427 | − 0.0714 | 0.04111 | − 1.74 | 3.01 | 0.121 |
| x 5 (salt solution) | 0.0794 | 0.0397 | 0.04111 | 0.97 | 0.93 | 0.362 |
| x 6 (KH2PO4) | − 0.2547 | − 0.1274 | 0.04111 | − 3.10 | 9.60 | 0.015* |
| x 7 (MgSO4.7H2O) | 0.3837 | 0.1918 | 0.04111 | 4.67 | 21.78 | 0.002** |
| x 8 (citric acid) | 0.0332 | 0.0166 | 0.04111 | 0.40 | 0.16 | 0.697 |
Values of “Prob. > F” less than 0.0010, 0.0100, and 0.0500 indicate that model terms are extremely significant (***), highly significant (**), and significant (*), respectively
Optimization of culture medium with BBD for L-Met production
Prior to modification the medium nutrition composition with BBD, the steepest ascent experiment was conducted (Suppl. Table S2), and the results suggested that the center point of the four significant components was chosen as 38 g/L glucose, 1.0 g/L yeast extract, 2.5 g/L KH2PO4, and 4.5 g/L MgSO4.7H2O. A further optimization regarding individual and interactive impacts of the four components on L-Met yield with more accuracy was implemented with a four-factor, three-level BBD, based on the sparsity-of-effects principle (Sibanda and Pretorius 2012). The design matrix and corresponding results were presented in Suppl. Table S3. The relationship between L-Met titer and variables was statistically evaluated by the F test for the ANOVA (Table 2), which represented the significance of the regression coefficient together with the p value. The model is extremely significant with a F value of 63.80 (p value of < 0.0001). The “Coefficient of Determination” (R2) value of the study was 0.9846, suggesting a high degree of correlation between the predicted values with BBD model and experimental values (Zou et al. 2015). In addition, the model “Pred R-Squared” of 0.9318 was in reasonable agreement with the “Adj R-Squared” of 0.9691. Thus, it could be applied to predict theoretical L-Met titer. “Adeq precision” measures the signal-to-noise ratio and a ratio greater than 4.00 is desirable. In the present work, the ratio of 24.911 indicates an adequate signal and suggests that this model could be used to navigate the design space. A low value of coefficient of variance (C.V.%) (5.59%) indicated a high degree of precision and reliability of the experimental values.
Table 2.
ANOVA analysis of BBD experiments
| Source | df | Sum of squares | Mean square | F value | p value prob.> F | Significant |
|---|---|---|---|---|---|---|
| Model | 14 | 7.88 | 0.56 | 63.80 | < 0.0001 | *** |
| x 1 (glucose) | 1 | 0.24 | 0.24 | 27.56 | 0.0001 | *** |
| x 3 (yeast extract) | 1 | 0.23 | 0.23 | 25.63 | 0.0002 | *** |
| x 6 (KH2PO4) | 1 | 2.12 | 2.12 | 239.93 | < 0.0001 | *** |
| x 7 (MgSO4.7H2O) | 1 | 0.042 | 0.042 | 4.75 | 0.0469 | * |
| x 1 x 3 | 1 | 6.999E-003 | 6.999E-003 | 0.79 | 0.3883 | |
| x 1 x 6 | 1 | 0.017 | 0.017 | 1.87 | 0.1930 | |
| x 1 x 7 | 1 | 7.321E-003 | 7.321E-003 | 0.83 | 0.3779 | |
| x 3 x 6 | 1 | 0.043 | 0.043 | 4.81 | 0.0456 | * |
| x 3 x 7 | 1 | 5.273E-003 | 5.273E-003 | 0.60 | 0.4524 | |
| x 6 x 7 | 1 | 0.012 | 0.012 | 1.40 | 0.2567 | |
| x 1 2 | 1 | 2.42 | 2.42 | 273.31 | < 0.0001 | *** |
| x 3 2 | 1 | 1.43 | 1.43 | 161.99 | < 0.0001 | *** |
| x 6 2 | 1 | 1.29 | 1.29 | 145.88 | < 0.0001 | *** |
| x 7 2 | 1 | 2.77 | 2.77 | 313.22 | < 0.0001 | *** |
| Residual | 14 | 0.12 | 8.828E-003 | |||
| Lack of fit | 10 | 0.084 | 8.408E-003 | 0.85 | 0.6214 | |
| Pure error | 4 | 0.040 | 9.879E-003 | |||
| Cor total | 28 | 8.01 | ||||
| Std. dev | 0.094 | |||||
| Mean | 1.68 | |||||
| C.V. (%) | 5.59 | |||||
| PRESS | 0.55 | |||||
| R-squared | 0.9846 | |||||
| Adj R-squared | 0.9691 | |||||
| Pred R-squared | 0.9318 | |||||
| Adeq precision | 24.911 |
Values of “Prob. > F” less than 0.001, 0.01, and 0.05 indicate that model terms are extremely significant (***), highly significant (**), and significant (*), respectively
According to the significance test results (Table 2), the linear coefficients , , , and , the interaction coefficient , as well as the quadratic coefficient , , , and are all significant at a 5% significance level. Therefore, they are the most influential factors; while the variables , , ,, and are insignificant model terms (p > 0.05). These insignificant factors was excluded and the optimal second-order polynomial Eq. (2) was established to describe L-Met biosynthesis:
| 2 |
where Y is the predicted response, , , , and are the coded values for glucose, yeast extract, KH2PO4, and MgSO4.7H2O, respectively. The optimal conditions for maximum L-Met production, calculated by setting the partial derivatives of Eq. (2) to zero with respect to the corresponding variables, were as follows: glucose 37.43 g/L, yeast extract 0.95 g/L, KH2PO4 1.82 g/L, and MgSO4.7H2O 4.51 g/L. The maximum response value of L-Met titer was estimated as 2.79 g/L.
The 3-D response surface curves were plotted to better understand the relationships between the response Y (L-Met titer) and the independent variables. As shown in Fig. 1a–f, each response surface plot presented a clear peak and the maximum L-Met titer was predicted within the range tested, indicating that further increase of glucose, yeast extract, KH2PO4, and MgSO4.7H2O concentrations would not cause a further increase in L-Met biosynthesis. Figure 1a showed the effects of glucose and yeast extract on L-Met titer, with a constant concentration of KH2PO4 and MgSO4.7H2O (central point level). L-Met titer generally increased to a peak value with an increase in glucose or yeast extract concentration and then decreased when glucose or yeast extract concentration further increased beyond 38 g/L or 1.0 g/L, respectively. Similar results were also found in the effects on L-Met production against glucose and KH2PO4 (Fig. 1b), glucose and MgSO4.7H2O (Fig. 1c), yeast extract and KH2PO4 (Fig. 1d), yeast extract and MgSO4.7H2O (Fig. 1e), KH2PO4 and MgSO4.7H2O (Fig. 1f). Especially it was evident that a symmetrical mound shape with a coordinate axis was observed in Fig. 1d, and the p value (0.0456, ANOVA) was lower than 0.05, suggesting a significant effect on the interaction between yeast extract and KH2PO4 concentrations. Additionally, the variable of KH2PO4 concentration was relatively more significant than the yeast extract concentration on the L-Met titer. From Fig. 1a–d, we can see that small variations in glucose, yeast extract, KH2PO4 and MgSO4.7H2O concentrations could cause great changes on L-Met titer, indicating the variables of square interaction (, , , and ) were all significant on the L-Met titer, which were consistent with the results of the ANOVA.
Fig. 1.
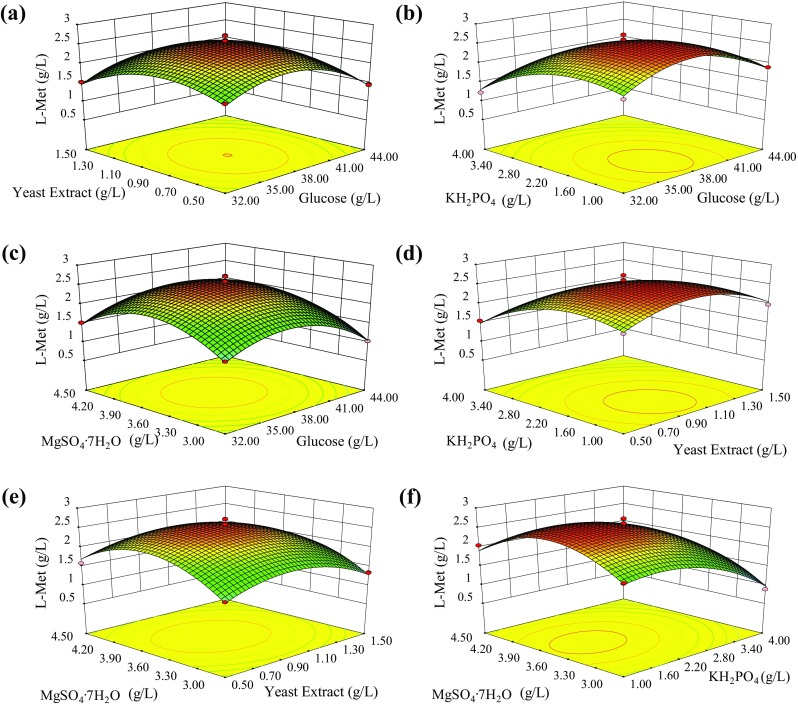
3-D response surface curves of the L-Met titer as a function of combinational effects of multiple variables. (a), glucose and yeast extract; (b), glucose and KH2PO4; (c), glucose and MgSO4.7H2O; (d), yeast extract and KH2PO4; (e), yeast extract and MgSO4.7H2O; (f), KH2PO4 and MgSO4.7H2O (f)
Experimental verification of theoretical optimum
In statistics, regression validation is a crucial phase of model analysis determining whether the numerical results quantifying hypothesized relationships between variables are acceptable as descriptions of the data (Giancristofaro and Salmaso 2007). The predicted results were validated by conducting solid experiments in triplicates. As expected, a mean experimental value of L-Met titer 3.04 ± 0.11 g/L was obtained, representing a good agreement with the predicted value (2.79 g/L). These results confirmed the validity and precision of the model, and the accumulation amount of L-Met was greatly increased compared with that when using the basal medium (< 2.0 g/L).
Batch fermentation for L-Met production in 5-L fermentor
Based on the shake flask results, the batch fermentation for L-Met production by E. coli MET-3 was carried out with the modified fermentation medium in 5-L fermentor. The results demonstrated that with the increase of glucose concentration in the fermentation medium, the L-Met yield against glucose was decreased (0.11 mol/mol at 20 g/L glucose versus 0.06 mol/mol at 40 g/L glucose), implying a possible catabolite repression effect induced by higher initial glucose concentration (Kim et al. 2004; Wan et al. 2011). Generally, this negative effect can be avoided by maintaining the glucose concentration at a lower level, and the demand for glucose can be achieved by glucose feeding in fed-batch process with an appropriate feeding mode (Kim et al. 2004). Herein, the glucose concentration was selected as 20 g/L to investigate the effects of other factors on both cell growth and L-Met biosynthesis. All experiments were repeated for three times. The optimal parameters would be adopted in the following fed-batch cultivation for further improvement of L-Met biosynthesis.
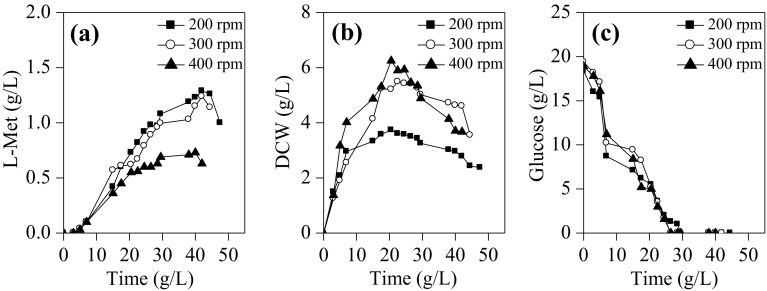
Agitation rate is an important factor influencing cell growth and metabolites biosynthesis. In this work, the agitation rates in a range of 200–400 rpm were tested. As shown in Fig. 2, glucose (20 g/L) was exhausted at approximately 25 h; and cell lysis occurred after 25 h, which might have been caused by the nutrition deficiency (Liu et al. 2017a). However, an increase of L-Met accumulation was still observed during glucose starvation until about 40 h. This was most likely due to the conversion from other components such as intermediate metabolites and byproducts in the medium. As shown in Fig. 2b, the maximum DCW of E. coli MET-3 at agitation rate of 200, 300, and 400 rpm was 3.7, 5.5, and 6.2 g/L, respectively, indicating that higher agitation rate (400 rpm) was favor to cell growth. On the contrary, higher L-Met titer (about 1.30 g/L) was achieved at lower agitation rate (200–300 rpm) (Fig. 2a). Thus, comprehensively considering cell growth and L-Met production, the optimal agitation rate was recommended as 300 rpm in batch fermentation.
Fig. 2.
The effect of different agitation rates on L-Met production by E. coli MET-3 during batch cultivation in 5-L fermentor. The medium pH was 7.0, and induction temperature was 28 °C. a L-Met biosynthesis; (b) cell growth; (c) glucose consumption. Data are the average of three batch cultures. For clarity, error bars are not displayed
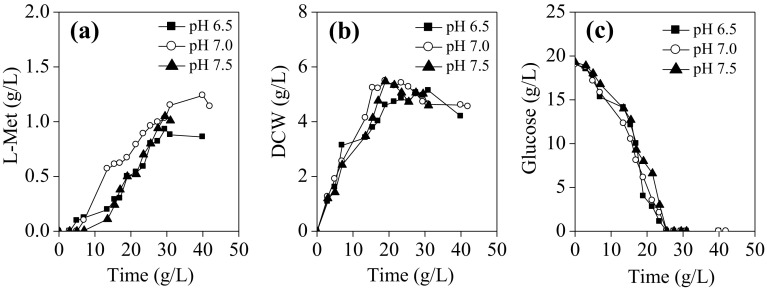
A pH range from 6.5 to 7.5 was tested for L-Met production. This pH range can assure high bioavailability of most nutrients essential for bacteria growth and metabolism. Figure 3 showed that, there were no significant differences in glucose consumption and biomass formation profiles for recombinant E. coli MET-3 at different medium pH tested. However, when pH was maintained at 7.0, the L-Met production was highest, which was 37.78% and 18.10% higher than that at pH 6.5 and 7.5, respectively; indicating that neutral pH (pH 7.0) was helpful to provide E. coli MET-3 cells a more conducive metabolic environment for L-Met biosynthesis.
Fig. 3.
The effect of different medium pH on L-Met production by E. coli MET-3 during batch cultivation in 5-L fermentor. The agitation rate was 300 rpm, and the induction temperature was 28 °C. a L-Met biosynthesis; (b) cell growth; (c) glucose consumption. Data are the average of three batch cultures. For clarity, error bars are not displayed
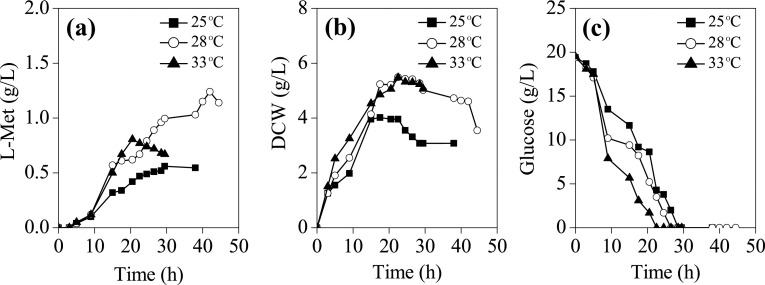
Temperature influences molecule movements and thus affects the mass transfer process and protonation of active sites of the enzymes (Liu et al. 2018a, b). Particularly, induction temperature is one of the major factors affecting protein expression levels in E. coli (Wang et al. 2018). Generally, higher induction temperature can lead to a higher protein expression rate and larger expression amount (Duan et al. 2017; Zheng et al. 2017). However, rapid protein generation will increase the chances of peptide misfolding and inclusion body formation (Duan et al. 2017). To avoid the insoluble expression of the essential enzymes in L-Met biosynthesis pathway after being induced, the cultures were subsequently incubated at relatively low temperatures (25 °C, 28 °C, and 33 °C). As shown in Fig. 4, with induction at 33 °C, the rate of glucose uptake, cell growth, and L-Met yielding prior to glucose depletion (at about 20 h) were apparently more rapid, compared with those at 25 °C and 28 °C. However, a dramatic decline in L-Met titer immediately followed after glucose limitation, which might due to a strengthened L-Met catabolism after glucose depletion through triggering re-orientation of metabolic pathways of E. coli MET-3. In contrast, at 25 °C, the metabolism rate was obviously decreased, reflected by the slower glucose utilization, biomass formation, and product accumulation. A moderate temperature (28 °C) resulted in an extension of fermentation period and significant increase in L-Met production. The maximum L-Met titer (1.24 g/L) was 1.09- and 2.21-fold higher than those at 33 °C and 25 °C, respectively. The results indicated that the activities of key enzymes participating in L-Met biosynthesis were enhanced at 28 °C, ensuring a strengthened L-Met synthesis by the E. coli cells.
Fig. 4.
The effect of different induction temperatures on L-Met production by E. coli MET-3 during batch cultivation in 5-L fermentor. The agitation rate was 300 rpm, and the medium pH was 7.0. a L-Met biosynthesis; (b) cell growth; (c) glucose consumption. Data are the average of three batch cultures. For clarity, error bars are not displayed
Enhanced L-Met production by fed-batch fermentation
To further characterize the fermentation properties of E. coli MET-3, fed-batch cultivation was conducted in a 5-L fermentor with the modified fermentation medium containing 20 g/L initial glucose under optimized cultivation conditions. During the fermentation process, the medium pH was maintained at 7.0 ± 0.2. The agitation rate was set at 300 rpm in the batch period and then adjusted between 200 and 900 rpm to keep the DO above 20%. The temperature was maintained at 37 °C until the OD600 reached about 2.0, after which it was lowered to 28 °C. After the initial glucose of 20 g/L was exhausted, supplementation of feeding medium was started immediately to maintain the residual glucose at 1–10 g/L.
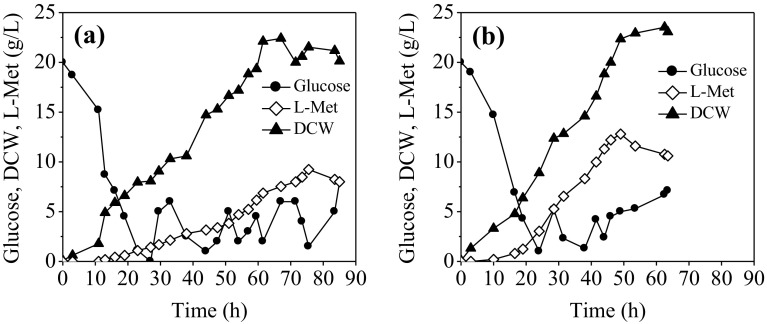
As shown in Fig. 5a, with the original fermentation medium, the DCW of E. coli MET-3 increased continuously and finally reached the maximum value of 22.42 g/L at 67 h, which was then slightly declined till the end of fermentation. After induction, L-Met accumulated gradually with a maximum titer of 9.24 g/L at 75.5 h. After then, L-Met yield dropped dramatically, which may be explained by the re-utilization of L-Met by other metabolic pathways at the late period of fermentation. When cultivation of E. coli MET-3 in the modified medium, the maximum DCW of 23.53 g/L and the maximum L-Met titer of 12.80 g/L was observed at 62.5 h and 49 h, respectively; which was increased by 38.53% with the productivity of 0.261 g/L/h, increased by 1.13-fold. Additionally, the L-Met yield against glucose reached 0.13 mol/mol, representing a 30% higher than the control. The results indicated that a more rapid cell growth and enhanced L-Met biosynthesis was achieved through using the modified fermentation medium in fed-batch fermentation. Although L-Met titer was still lower than the highest level (35 g/L) reported so far (Li et al. 2017; Willke 2014), it is worth noting that, during the fermentation process, several organic acids such as acetic acid, formic acid, and pyruvic acid, as well as other amino acids such as valine, homoserine, and leucine were also detected (data not shown), which competitively reduce the carbon flux distribution into L-Met biosynthesis. Therefore, it is believed that further modification of E. coli strain MET-3 by blocking the byproduct formation pathway, together with adoption of a more suitable fed-batch fermentation would result in greater enhancement of L-Met production.
Fig. 5.
Comparison of the L-Met production by E. coli MET-3 with original fermentation medium (a) and modified fermentation medium (b) during fed-batch fermentation in 5-L fermentor. Data are the average of three batch cultures. For clarity, error bars are not displayed
Conclusions
In this study, the fermentation conditions for genetically engineered E. coli MET-3 was optimized, the present study was undertaken to design the medium with the aids of statistical methodology PB design and BBD and to optimize fermentation conditions to improve the L-Met production. According to the statistical methodology PB and BBD design, the concentration of the most important medium components were determined to be as follows: glucose 37.43 g/L, yeast extract 0.95 g/L, KH2PO4 1.82 g/L, and MgSO4.7H2O 4.51 g/L. Under which the L-Met titer was increased to 3.04 ± 0.11 g/L from less than 2.0 g/L. When performed in a batch fermentation in 5-L fermentor at optimized fermentation medium and cultivation conditions with agitation rate of 300 rpm, medium pH of 7.0, and induction temperature of 28 °C, a maximum L-Met yield of 12.80 g/L was obtained, which was increased by 38.53% with a yield against glucose (mol/mol) increased by 30%. Moreover, a 2.13-fold higher productivity (0.261 g/L/h) was achieved in comparison to the original medium. This result confirmed that the designed medium and optimized fermentation conditions were beneficial for L-Met fermentative production.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors gratefully acknowledge the financial support of the National Natural Science Foundation of China (No. 31500031), the Natural Science Foundation of Zhejiang Province (No. LQ14B060004), and the Natural Science Foundation of Zhejiang University of Technology (No. 2012XY010).
Conflict of interest
The authors declared that they have no competing interests.
Ethical statement
This article does not contain any studies with human participants or animals performed by any of the authors.
References
- Asadi N, Zilouei H. Optimization of organosolv pretreatment of rice straw for enhanced biohydrogen production using Enterobacter aerogenes. Bioresour Technol. 2017;227:335–344. doi: 10.1016/j.biortech.2016.12.073. [DOI] [PubMed] [Google Scholar]
- Bicas JL, Barros FFC, Wagner R, Godoy HT, Pastore GM. Optimization of R-(+)-α-terpineol production by the biotransformation of R-(+)-limonene. J Ind Microbiol Biotechnol. 2008;35:1061–1070. doi: 10.1007/s10295-008-0383-0. [DOI] [PubMed] [Google Scholar]
- Chen Y, Li D, Dai Z, Piao X, Wu Z, Wang B, Zhu Y, Zeng Z. L-Methionine supplementation maintains the integrity and barrier function of the small-intestinal mucosa in post-weaning piglets. Amino Acids. 2014;46:1131–1142. doi: 10.1007/s00726-014-1675-5. [DOI] [PubMed] [Google Scholar]
- Dayana PS, Bakthavatsalam AK. Optimization of phenol degradation by the microalga Chlorella pyrenoidosa using Plackett–Burman design and response surface methodology. Bioresour Technol. 2016;207:150–156. doi: 10.1016/j.biortech.2016.01.138. [DOI] [PubMed] [Google Scholar]
- Dischert W, Vasseur P, Figge R (2016) Microorganism for methionine production with improved methionine synthase activity and methionine efflux. United States Patent (US20160177352A1)
- Duan X, Hu S, Qi X, Gu Z, Wu J. Optimal extracellular production of recombinant Bacillus circulans β-galactosidase in Escherichia coli BL21(DE3) Process Biochem. 2017;53:17–24. doi: 10.1016/j.procbio.2016.11.008. [DOI] [Google Scholar]
- Figge R, Soucaille P, Barbier G, Bestel-Corre G, Boisart C, Chateau M (2016) Increasing methionine yield. European Patent (EP2573189:B1)
- Giancristofaro RA, Salmaso L. Model performance analysis and model validation in logistic regression. Statistica. 2007;63:375–396. doi: 10.6092/issn.1973-2201/358. [DOI] [Google Scholar]
- Gomes J, Kumar D. Production of L-methionine by submerged fermentation: a review. Enzyme Microb Technol. 2005;37:3–18. doi: 10.1016/j.enzmictec.2005.02.008. [DOI] [Google Scholar]
- Hou FN, Mu TH, Ma MM, Blecker C. Optimization of processing technology using response surface methodology and physicochemical properties of roasted sweet potato. Food Chem. 2019;278:136–143. doi: 10.1016/j.foodchem.2018.11.034. [DOI] [PubMed] [Google Scholar]
- Huang JF, Liu ZQ, Jin LQ, Tang XL, Shen ZY, Yin HH, Zheng YG. Metabolic engineering of Escherichia coli for microbial production of L-methionine. Biotechnol Bioeng. 2017;114:843–851. doi: 10.1002/bit.26198. [DOI] [PubMed] [Google Scholar]
- Huang JF, Shen ZY, Mao QL, Zhang XM, Zhang B, Wu JS, Liu ZQ, Zheng YG. Systematic analysis of bottlenecks in a multibranched and multilevel regulated pathway: the molecular fundamentals of L-methionine biosynthesis in Escherichia coli. ACS Syn Biol. 2018;7:2577–2589. doi: 10.1021/acssynbio.8b00249. [DOI] [PubMed] [Google Scholar]
- Huang JF, Zhang B, Shen ZY, Liu ZQ, Zheng YG. Metabolic engineering of E. coli for the production of O-succinyl-L-homoserine with high yield. 3 Biotech. 2018;8:310. doi: 10.1007/s13205-018-1332-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowski J, Kubińska M, Zduńczyk Z. Nutritional and immunomodulatory function of methionine in poultry diets-a review. Ann Anim Sci. 2014;14:17–31. doi: 10.2478/aoas-2013-0081. [DOI] [Google Scholar]
- Kase H, Nakayama K. L-Methionine production by methionine analog-resistant mutants of Corynebacterium glutamicum. Agr Biol Chem. 1975;39:153–160. doi: 10.1271/bbb1961.39.153. [DOI] [Google Scholar]
- Kim YK, Kang DG, Choi SS, Kim JH, Chung JC, Cha HJ. Co-expression of bacterial hemoglobin overrides high glucose-induced repression of foreign protein expression in Escherichia coli W3110. Biotechnol Lett. 2004;26:1173–1178. doi: 10.1023/B:BILE.0000035491.64912.84. [DOI] [PubMed] [Google Scholar]
- Kumar D, Garg S, Bisaria VS, Sreekrishnan TR, Gomes J. Production of methionine by a multi-analogue resistant mutant of Corynebacterium lilium. Process Biochem. 2003;38:1165–1171. doi: 10.1016/S0032-9592(02)00287-X. [DOI] [Google Scholar]
- Labbeiki G, Attar H, Heydarinasab A, Sorkhabadi S, Rashidi A. Enhanced oxygen transfer rate and bioprocess yield by using magnetite nanoparticles in fermentation media of erythromycin. DARU J Pharm Sci. 2014;22:66. doi: 10.1186/s40199-014-0066-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A, Choi KH, Yoon D, Kim S, Cha J. Characterization of a thermostable glycoside hydrolase family 36 alpha-galactosidase from Caldicellulosiruptor bescii. J Biosci Bioeng. 2017;124:289–295. doi: 10.1016/j.jbiosc.2017.04.011. [DOI] [PubMed] [Google Scholar]
- Li H, Wang BS, Li YR, Zhang L, Ding ZY, Gu ZH, Shi GY. Metabolic engineering of Escherichia coli W3110 for the production of L-methionine. J Ind Microbiol Biotechnol. 2017;44:75–88. doi: 10.1007/s10295-016-1870-3. [DOI] [PubMed] [Google Scholar]
- Liu ZQ, Dong SC, Yin HH, Xue YP, Tang XL, Zhang XJ, He JY, Zheng YG. Enzymatic synthesis of an ezetimibe intermediate using carbonyl reductase coupled with glucose dehydrogenase in an aqueous-organic solvent system. Bioresour Technol. 2017;229:26–32. doi: 10.1016/j.biortech.2016.12.098. [DOI] [PubMed] [Google Scholar]
- Liu ZQ, Wu L, Zhang XJ, Xue YP, Zheng YG. Directed evolution of carbonyl reductase from Rhodosporidium toruloides and its application in stereoselective synthesis of tert-butyl (3R,5S)-6-chloro-3,5-dihydroxyhexanoate. J Agr Food Chem. 2017;65:3721–3729. doi: 10.1021/acs.jafc.7b00866. [DOI] [PubMed] [Google Scholar]
- Liu ZQ, Lu MM, Zhang XH, Cheng F, Xu JM, Xue YP, Jin LQ, Wang YS, Zheng YG. Significant improvement of the nitrilase activity by semi-rational protein engineering and its application in the production of iminodiacetic acid. Int J Biol Macromol. 2018;116:563–571. doi: 10.1016/j.ijbiomac.2018.05.045. [DOI] [PubMed] [Google Scholar]
- Liu ZQ, Wu L, Zheng L, Wang WZ, Zhang XJ, Jin LQ, Zheng YG. Biosynthesis of tert-butyl (3R,5S)-6-chloro-3,5-dihydroxyhexanoate by carbonyl reductase from Rhodosporidium toruloides in mono and biphasic media. Bioresour Technol. 2018;249:161–167. doi: 10.1016/j.biortech.2017.09.204. [DOI] [PubMed] [Google Scholar]
- Ma L, Wang L, Tang J, Yang Z. Optimization of arsenic extraction in rice samples by Plackett–Burman design and response surface methodology. Food Chem. 2016;204:283–288. doi: 10.1016/j.foodchem.2016.02.126. [DOI] [PubMed] [Google Scholar]
- Mampel J, Schröder H, Haefner S, Sauer U. Single-gene knockout of a novel regulatory element confers ethionine resistance and elevates methionine production in Corynebacterium glutamicum. Appl Microbiol Biotechnol. 2005;68:228–236. doi: 10.1007/s00253-005-1893-6. [DOI] [PubMed] [Google Scholar]
- Maresi E, Janson G, Fruncillo S, Paiardini A, Vallone R, Dominici P, Astegno A. Functional characterization and structure-guided mutational analysis of the transsulfuration enzyme cystathionine-lyase from Toxoplasma gondii. Int J Mol Sci. 2015;19:1–19. doi: 10.3390/ijms19072111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem. 1959;31:426–428. doi: 10.1021/ac60147a030. [DOI] [Google Scholar]
- Mondal S, Das YB, Chatterjee SP. Methionine production by microorganisms. Folia Microbiol. 1996;41:465–472. doi: 10.1007/BF02814659. [DOI] [PubMed] [Google Scholar]
- Park SD, Lee JY, Sim SY, Kim Y, Lee HS. Characteristics of methionine production by an engineered Corynebacterium glutamicum strain. Metab Eng. 2007;9:327–336. doi: 10.1016/j.ymben.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Plackett RL, Burman JP. The design of optimum multifactorial experiments. Biometrika. 1946;33:305–325. doi: 10.2307/2332195. [DOI] [Google Scholar]
- Rahman Z, Zidan AS, Habib MJ, Khan MA. Understanding the quality of protein loaded PLGA nanoparticles variability by Plackett–Burman design. Int J Pharm. 2010;389:186–194. doi: 10.1016/j.ijpharm.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazmohammad H, Omidinia E. Medium optimization for improved production of dihydrolipohyl dehydrogenase from Bacillus sphaericus PAD-91 in Escherichia coli. Mol Biotechnol. 2017;59:260–270. doi: 10.1007/s12033-017-0013-z. [DOI] [PubMed] [Google Scholar]
- Sibanda W, Pretorius P. Comparative study of the application Box Behnken design (BBD) and binary logistic regression (BLR) to study the effect of demographic characteristics on HIV risk in south Africa. JAMS. 2012;1:15–40. doi: 10.1007/s13721-013-0032-z. [DOI] [Google Scholar]
- Teleki A, Rahnert M, Bungart O, Gann B, Ochrombel I, Takors R. Robust identification of metabolic control for microbial L-methionine production following an easy-to-use puristic approach. Metab Eng. 2017;41:159–172. doi: 10.1016/j.ymben.2017.03.008. [DOI] [PubMed] [Google Scholar]
- Usuda Y, Kurahashi O. Effects of deregulation of methionine biosynthesis on methionine excretion in Escherichia coli. Appl Environ Microbiol. 2005;71:3228–3234. doi: 10.1128/AEM.71.6.3228-3234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan M, Liu P, Xia J, Rosenberg JN, Oyler GA, Betenbaugh MJ, Nie Z, Qiu G. The effect of mixotrophy on microalgal growth, lipid content, and expression levels of three pathway genes in Chlorella sorokiniana. Appl Microbiol Biotechnol. 2011;91:835–844. doi: 10.1007/s00253-011-3399-8. [DOI] [PubMed] [Google Scholar]
- Wang XX, Lin CP, Zhang XJ, Liu ZQ, Zheng YG. Improvement of a newly cloned carbonyl reductase and its application to biosynthesize chiral intermediate of duloxetine. Process Biochem. 2018;70:124–128. doi: 10.1016/j.procbio.2018.04.010. [DOI] [Google Scholar]
- Weissbach H, Brot N. Regulation of methionine synthesis in Escherichia coli. Mol Microbiol. 1991;5:1593–1597. doi: 10.1111/j.1365-2958.1991.tb01905.x. [DOI] [PubMed] [Google Scholar]
- Willke T. Methionine production-a critical review. Appl Microbiol Biotechnol. 2014;98:9893–9914. doi: 10.1007/s00253-014-6156-y. [DOI] [PubMed] [Google Scholar]
- Ye Z, Wang W, Yuan Q, Ye H, Sun Y, Zhang H, Zeng X. Box–Behnken design for extraction optimization, characterization and in vitro antioxidant activity of Cicer arietinum L. hull polysaccharides. Carbohydr Polym. 2016;147:354–364. doi: 10.1016/j.carbpol.2016.03.092. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhao T, Cheng T, Liu X, Zhang H. Rapid resolution liquid chromatography (RRLC) analysis of amino acids using pre-column derivatization. J Chromatogr B Analyt Technol Biomed Life Sci. 2012;906:91–95. doi: 10.1016/j.jchromb.2012.08.030. [DOI] [PubMed] [Google Scholar]
- Zheng YG, Yin HH, Yu DF, Chen X, Tang XL, Zhang XJ, Xue YP, Wang YJ, Liu ZQ. Recent advances in biotechnological applications of alcohol dehydrogenases. Appl Microbiol Biotechnol. 2017;101:987–1001. doi: 10.1007/s00253-016-8083-6. [DOI] [PubMed] [Google Scholar]
- Zou SP, Zhong W, Xia CJ, Gu YN, Niu K, Zheng YG, Shen YC. Mutagenesis breeding of high echinocandin B producing strain and further titer improvement with culture medium optimization. Bioprocess Biosyst Eng. 2015;38:1845–1854. doi: 10.1007/s00449-015-1425-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.