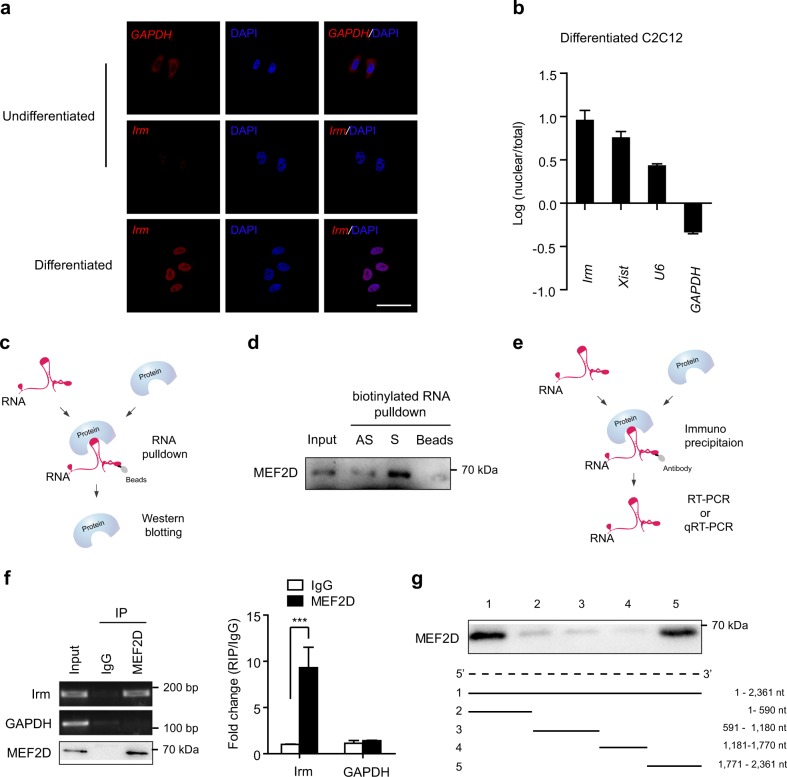
Fig. 5. Irm directly binds to MEF2D.
a RNA-FISH for detecting Irm and GAPDH in undifferentiated and differentiated C2C12 cells. Red: Irm or GAPDH. Blue: DAPI staining. Scale bar, 50 μm. b Relative abundance of Irm in total and nuclear RNAs of differentiating C2C12 cells, as detected by qRT-PCR. U6, Xist, and GAPDH were used as endogenous controls. c A schematic representation of RNA pull-down assay. d Western blotting assay for the specific interaction of Irm with MEF2D. e A schematic representation of RNA immunoprecipitation (RIP) assay. f RIP assay showed the association of MEF2D with Irm in vivo, as detected by RT-PCR and qRT-PCR. g Deletion mapping of the MEF2D-binding region(s) in Irm. Top: western blotting for MEF2D in protein samples pulled down by the different truncated Irm constructs. Bottom: diagrams of the full-length and truncated Irms. nt: nucleotide. Data shown represent the mean ± SEM of three independent experiments. n.s., not significant; ***P < 0.05 by Student’s t test. DAPI, 4′,6-diamidino-2-phenylindole; qRT-PCR, quantitative real-time polymerase chain reaction; RNA-FISH, RNA fluorescence in situ hybridization