Abstract
Cytokine gene single nucleotide polymorphisms (SNPs) can influence cytokine levels, which may be associated with tuberculosis (TB) susceptibility. There is evidence that interleukin 1B (IL1B), tumor necrosis factor-alpha (TNF-alpha), and IL6 may be involved in the progression of TB. Using a self-validating case-control design, we selected eleven functional SNPs in IL1B, TNF and IL6 to detect their association with TB in Chinese Han and Tibetan populations. The associations between SNPs and TB were estimated by computing the odds ratios (ORs) and 95% confidence intervals (95% CI) using logistic regression analyses. We found that the IL1B rs16944 polymorphism was associated with decreased risk of TB in the two studies. The G allele at rs2069837 of IL6 was significantly more common in controls than in TB patients in the Han population. Moreover, TNF rs1799964 and rs1800630 were risk factors for susceptibility to TB, which were validated in the Chinese Tibetan population. In addition, TNF rs1799724 and rs1800629 were associated with TB, but only in the Tibetan population. In conclusion, SNPs of the IL1B and TNF gene were associated with TB susceptibility in Chinese Han and Tibetan populations. IL6 polymorphism may be considered as a protective factor for TB in the Chinese Han population, but not the Tibetan population.
Introduction
Tuberculosis (TB) is an ancient infectious disease caused by Mycobacterium tuberculosis (M. TB) which can spread not only to the respiratory system but also to other organs. In 2017, the World Health Organization (WHO) reported that the incidence of TB in China was 64 per 100,000 individuals1. China has ranked the number three regarding the number of TB patients and the Tibetan nationality in this country has the highest incidence rate2. An estimated one-third of the world’s population is infected with M. TB, but only 5–15% of infected individuals develop active TB and the rest remain asymptomatic3. The reason why only a minority of the infected individuals develops TB disease is still largely unknown. It was suggested that the interactions between the bacterial agent, environmental and genetic factors play important roles in the progression to TB disease4.
Studies of concordance of disease in twins have shown that host genetic factors play significant roles in TB disease5,6. Determining the specific host genes associated with TB disease may enhance the understanding of the pathogenesis of TB and further the development of treatment strategies. To date, many studies have suggested that cytokine gene polymorphisms are associated with TB among different populations. For example, interferon gamma (IFNG) and interleukin 17 (IL17) polymorphisms were reported to be associated with TB risk7,8. Several meta-analyses suggested that cytokines such as IL10, IL12B, IL27 and CC chemokine ligand 5 (CCL5) play important roles in the progression of TB9–12. Indeed, genetic variants in cytokine genes could affect the recognition sites of the transcription factors, leading to altered transcriptional activity, which may then result in a change in cytokine production levels13. Intricate interplay among lymphocytes, antigen-presenting cells and released cytokines is involved in the immune response against TB.
Polymorphisms in the IL1B, tumor necrosis factor (TNF), and IL6 genes have been associated with susceptibility to TB in previous studies. The IL1B gene is located on chromosome 2. IL1B is a major proinflammatory cytokine that is produced by monocytes, macrophages and dendritic cells during infection and inflammation. IL1B gene polymorphisms have been demonstrated alter IL1B protein production14,15. IL1B plays a critical role in the immune response to mycobacteria, and may increase resistance to initial infection. IL1B polymorphisms has been associated with TB in human16. TNF-alpha is a multifunctional cytokine that is synthesized by monocytes, macrophages, neutrophils, T-cells, and natural killer-cells17. These cells have been described to play a critical role in the immunopathogenesis of TB. TNF-alpha also participates in the granulomatous reaction which could further limit M. TB growth. Single nucleotide polymorphisms (SNPs) within TNF were suggested substantially to affect TNF-alpha production levels18. Several studies have shown a significant association between TNF polymorphisms and TB19,20. IL6 is produced by activated monocytes and macrophages. It is an important immunoregulatory factor, which can reduce the production of IL1B and TNF-alpha21. Previous studies have shown that TB patients had higher IL6 levels, compared with healthy controls22. Also, it could increase early IFN-γ secretion in MTB infection and thus help to control TB infection23. IL6-deficient mice were more susceptible to TB than wild-type mice24 and polymorphisms in IL6 were reported to be associated with TB25.
Despite the wealth of association studies of IL1B, TNF and IL6 polymorphisms a consensus has yet to emerge as to which variants affect susceptibility to TB. In addition, none of the previous studies was conducted with a validation cohort. Thus, we hypothesized that ILB, IL6 and TNF polymorphisms are associated with TB in Chinese Han and Tibetan populations, and performed two independent studies with TB cases and healthy controls in these two populations to determine the influence of the three cytokine gene polymorphisms on TB susceptibility.
Results
Demographics of the study population
In the initial cohort, we recruited 636 TB cases (50.9% males) and 608 controls (49.7% males) from the Chinese Han population. The mean (±SD) age was 36.8 (±15.7) years for cases and 37.1 (±15.7) years for controls. There were no significant differences in sex and age between groups (Table 1). The percentage of smoking is lower in cases than in controls (P = 0.003) (Table 1). In the validation cohort, 613 TB patients (53.3% males; mean (±SD) age was 34.5 (±14.5) years) and 603 healthy subjects (55.2% males; mean (±SD) age was 34.6 (±13.8) years) were enrolled from the Tibetan population. Cases and controls in this group were also well-matched by age and gender (Table 1). However, data of smoking status among the Tibetan subjects was not collected because smoking rates were very low in this population for cultural reasons.
Table 1.
Demographic distribution of healthy controls and tuberculosis patients.
| Parameters | Cases | Controls | P value |
|---|---|---|---|
| Han population | n = 636 | n = 608 | |
| Age (mean ± SD, years) | 36.8 ± 15.7 | 37.1 ± 15.7 | 0.677 |
| Male, n (%) | 324 (50.9) | 302 (49.7) | 0.654 |
| Smoking, n (%) | 195 (69.3) | 141 (76.8) | 0.003? |
| Location of TB, n (PTB/EPTB) | 276/360 | ||
| Acid-fast bacilli stain positive, n (positive/negative) | 138/360 | ||
| Culture positive n (positive/negative) | 32/126 | ||
| TB-DNA positive n (positive/negative) | 122/133 | ||
| Tibetan population | n = 613 | n = 603 | |
| Age, (mean ± SD, years) | 34.5 ± 14.5 | 34.6 ± 13.8 | 0.909 |
| Male, n (%) | 327 (53.3) | 333 (55.2) | 0.511 |
Abbreviations: SD, standard deviation; PTB, pulmonary tuberculosis; EPTB, extra-pulmonary tuberculosis.
Association between SNPs and TB susceptibility in two studies
Eleven SNPs previously investigated in genetic studies of TB in the IL1B, TNF and IL6 genes were selected for this study (Table S1 and Table S2). Among the 11 SNPs, rs114362326, rs206983727, rs179972428, rs180062928, and rs180063028 were associated with transcriptional activity of the genes. rs36152529, rs179972430, rs180063031, rs180062932, rs179996431, rs114363433, rs1694434, and rs180079535 polymorphisms may influence protein levels of their respective cytokines. rs180063036, rs179972436, rs180079537, rs1694438, rs180062938, rs179996439 and rs36152540 were contributory factors to TNF gene expression. rs17147230 was predicted to have functional significance by the FuncPred algorithm. Only rs1800630 deviated from HWE and only in the Tibetan control subjects (P = 0.024). The characteristics of the SNPs are shown in Table 2.
Table 2.
Characteristics of the SNPs in the study.
| Gene/SNPs | Chr. | SNP | Location in gene | Function* | MA | MAF | MA | MAF | HWE | |
|---|---|---|---|---|---|---|---|---|---|---|
| Han | Han | Tibetan | Tibetan | Han | Tibetan | |||||
| IL1B | 2 | rs1143634 | exon5 (nonsynonymous) | IL1B levels | A | 0.02 | A | 0.03 | 0.340 | 0.776 |
| rs16944 | 5′FLANKING | IL1B expression, IL1B levels | A | 0.48 | G | 0.42 | 0.999 | 0.976 | ||
| rs1143623 | 5′FLANKING | TFBS | G | 0.41 | G | 0.48 | 0.731 | 0.991 | ||
| IL6 | 7 | rs17147230 | Unkonwn | TFBS | T | 0.44 | A | 0.48 | 0.645 | 0.554 |
| rs1800795 | 5′FLANKING | IL6 expression, IL6 levels | C | 0.002 | C | 0.002 | 0.993 | 0.999 | ||
| rs2069837 | intron2 | TFBS | C | 0.17 | C | 0.26 | 0.366 | 0.118 | ||
| TNF | 6 | rs1799964 | 5′FLANKING | TNF expression, TNF levels | C | 0.17 | C | 0.22 | 0.986 | 0.075 |
| rs1800630 | 5′FLANKING | TNF expression, TFBS, TNF-alpha levels | A | 0.16 | A | 0.18 | 0.841 | 0.024 | ||
| rs1799724 | 5′FLANKING | TNF expression, TFBS, TNF-alpha levels | C | 0.14 | C | 0.16 | 0.951 | 0.989 | ||
| rs1800629 | 5′FLANKING | TNF expression, TNF-alpha levels | A | 0.07 | A | 0.02 | 0.869 | 0.092 | ||
| rs361525 | 5′FLANKING | TNF expression, TNF-alpha levels | A | 0.02 | A | 0.04 | 0.281 | 0.686 | ||
Abbreviation: SNP, single nucleotide polymorphism; MA, minor allele; MAF, minor allele frequency; HWE, Hardy Weinberg equilibrium; TFBS, transcription factor binding sites; Chr. chromosome.
*Function of each SNP was reported by previous studies and/or predicted from the NIH FuncPred website (https://snpinfo.niehs.nih.gov/snpinfo/snpfunc.html).
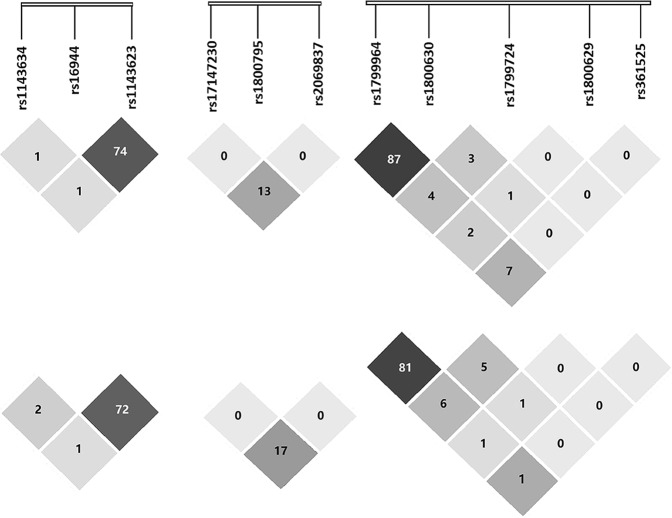
We observed that the three gene polymorphisms were associated with TB (Tables 3 and 4). In IL1B, at the rs1143634 polymorphic site, the AA + GA genotypes compared with GG were protective factors against TB (P = 0.042). In IL6, the rs2069837 G allele (P = 0.046) and the G carriers (GG + GA) (P = 0.044) were associated with decreased risk of TB. In TNF, two SNPs were significantly associated with TB under the dominant model, i.e., rs1799964 (P = 0.017) and rs1800630 (P = 0.002). We also determined that CT heterozygotes and the C allele of rs1799964 were risk factors for susceptibility to TB. In addition, CA heterozygotes (P = 0.024) and the A allele (P = 0.020) of rs1800630 occurred at a higher frequency in the TB cases than in the control group. The LD between the SNPs is shown in Fig. 1. We found that the TNF frequency of haplotype CACGG was significantly different between TB and control groups (Table 5).
Table 3.
Genotype distribution of cytokine genes in the two populations.
| Gene/SNPs | Han population | Tibetan population | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Case(%), n = 636 | Control(%), n = 608 | P # | OR#(95% CI) | Case(%), n = 613 | Control(%), n = 603 | P # | OR#(95% CI) | |||
| IL1B | rs1143634 | Genotype | ||||||||
| GG | 604 (95.6) | 288 (94.4) | 566 (92.3) | 568 (94.4) | ||||||
| GA | 28 (4.4) | 16 (5.2) | 0.094 | 1.71 (0.91–3.19) | 46 (7.5) | 34 (5.6) | 0.184 | 1.37 (0.86–2.16) | ||
| AA | 0 (0) | 1 (0.3) | — | 1 (0.2) | 0 (0.0) | — | ||||
| Allele | ||||||||||
| G | 1236 (97.8) | 592 (97.0) | 1178 (96.1) | 1170 (97.2) | ||||||
| A | 28 (2.2) | 18 (3.0) | 0.183 | 1.50 (0.83–2.73) | 48 (3.9) | 34 (2.8) | 0.133 | 1.41 (0.90–2.21) | ||
| rs16944 | Genotype | |||||||||
| GG | 164 (25.9) | 164 (27.1) | 110 (17.9) | 131 (21.7) | 0.028 | 0.69 (0.50–0.96) | ||||
| GA | 309 (48.9) | 302 (49.9) | 0.463 | 0.90 (0.68–1.19) | 298 (48.6) | 303 (50.2) | 0.105 | 0.81 (0.62–1.05) | ||
| AA | 159 (25.2) | 139 (23.0) | 0.413 | 0.88 (0.64–1.20) | 205 (33.4) | 169 (28.0) | ||||
| Allele | ||||||||||
| G | 637 (50.4) | 630 (52.1) | 518 (42.3) | 565 (46.8) | 0.023 | 0.83 (0.71–0.97) | ||||
| A | 627 (49.6) | 580 (47.9) | 0.422 | 1.07 (0.91–1.25) | 708 (57.7) | 641 (53.2) | ||||
| rs1143623 | Genotype | |||||||||
| CC | 198 (31.3) | 203 (33.6) | 155 (25.3) | 162 (26.9) | ||||||
| GC | 309 (48.9) | 303 (50.1) | 0.709 | 1.05 (0.82–1.35) | 294 (48.0) | 299 (49.7) | 0.731 | 1.05 (0.80–1.38) | ||
| GG | 125 (19.8) | 99 (16.4) | 0.132 | 1.29 (0.93–1.79) | 164 (26.8) | 141 (23.4) | 0.226 | 1.22 (0.89–1.67) | ||
| Allele | ||||||||||
| C | 705 (55.8) | 709 (58.6) | 604 (49.3) | 623 (51.7) | ||||||
| G | 559 (44.2) | 501 (41.4) | 0.163 | 1.12 (0.96–1.31) | 622 (50.7) | 581 (48.3) | 0.222 | 1.10 (0.94–1.30) | ||
| IL6 | rs17147230 | Genotype | ||||||||
| AA | 197 (31.2) | 183 (30.2) | 123 (20.1) | 143 (23.8) | 0.290 | 0.84 (0.61–1.16) | ||||
| AT | 315 (49.8) | 289 (47.8) | 0.214 | 1.21 (0.90–1.62) | 313 (51.1) | 287 (47.7) | 0.665 | 1.06 (0.81–1.38) | ||
| TT | 120 (19.0) | 133 (22.0) | 0.273 | 1.20 (0.87–1.65) | 177 (28.9) | 172 (28.6) | ||||
| Allele | ||||||||||
| A | 709 (56.1) | 655 (54.1) | 559 (45.6) | 573 (47.6) | 0.344 | 0.93 (0.79–1.09) | ||||
| T | 555 (43.9) | 555 (45.9) | 0.328 | 0.92 (0.79–1.08) | 667 (54.4) | 631 (52.4) | ||||
| rs1800795 | Genotype | |||||||||
| GG | 630 (99.7) | 599 (99.0) | 606 (98.9) | 601 (99.7) | ||||||
| GC | 2 (0.3) | 6 (1.0) | 0.161 | 0.32 (0.06–1.58) | 6 (1.0) | 2 (0.3) | 0.185 | 2.96 (0.60–14.74) | ||
| CC | 0 (0) | 0 (0) | — | 1 (0.2) | 0 (0) | — | ||||
| Allele | ||||||||||
| G | 1262 (99.8) | 1204 (99.5) | 1218 (99.3) | 1204 (99.8) | ||||||
| C | 2 (0.2) | 6 (0.5) | 0.161 | 0.32 (0.06–1.58) | 8 (0.7) | 2 (0.2) | 0.082 | 3.97 (0.84–18.74) | ||
| rs2069837 | Genotype | |||||||||
| AA | 443 (70.1) | 392 (64.8) | 340 (55.5) | 338 (56.1) | ||||||
| GA | 163 (25.8) | 183 (30.2) | 0.060 | 0.79 (0.61–1.01) | 226 (36.9) | 213 (35.4) | 0.674 | 1.05 (0.83–1.34) | ||
| GG | 26 (4.1) | 30 (5.0) | 0.328 | 0.76 (0.44–1.31) | 47 (7.7) | 51 (8.5) | 0.697 | 0.92 (0.60–1.41) | ||
| Allele | ||||||||||
| A | 1049 (83.0) | 967 (79.9) | 906 (73.9) | 889 (73.8) | ||||||
| G | 215 (17.0) | 243 (20.1) | 0.046 | 0.81 (0.66–0.99) | 320 (26.1) | 315 (26.2) | 0.972 | 0.99 (0.83–1.20) | ||
| TNF | rs1799964 | Genotype | ||||||||
| TT | 390 (61.7) | 413 (68.3) | 329 (53.7) | 372 (61.8) | ||||||
| CT | 212 (33.5) | 173 (28.6) | 0.037 | 1.30 (0.02–1.66) | 239 (39.0) | 190 (31.6) | 0.004 | 1.43 (1.12–1.82) | ||
| CC | 30 (4.7) | 19 (3.1) | 0.099 | 1.65 (0.91–2.98) | 45 (7.3) | 40 (6.6) | 0.286 | 1.28 (0.81–2.01) | ||
| Allele | ||||||||||
| T | 992 (78.5) | 999 (82.6) | 897 (73.2) | 934 (77.6) | ||||||
| C | 272 (21.5) | 211 (17.4) | 0.011 | 1.30 (1.06–1.58) | 329 (26.8) | 270 (22.4) | 0.011 | 1.27 (1.06–1.53) | ||
| rs1800630 | Genotype | |||||||||
| CC | 410 (64.9) | 431 (71.2) | 372 (60.7) | 417 (69.2) | ||||||
| CA | 199 (31.5) | 157 (26.0) | 0.024 | 1.33 (1.04–1.71) | 208 (33.9) | 157 (26.0) | 0.002 | 1.49 (1.16–1.92) | ||
| AA | 23 (3.6) | 17 (2.8) | 0.312 | 1.39 (0.73–2.65) | 33 (5.4) | 29 (4.8) | 0.359 | 1.28 (0.76–2.14) | ||
| Allele | ||||||||||
| C | 1019 (80.6) | 1019 (84.2) | 952 (77.7) | 991 (82.2) | ||||||
| A | 245 (19.4) | 191 (15.8) | 0.020 | 1.28 (1.04–1.58) | 274 (22.3) | 215 (17.8) | 0.005 | 1.33 (1.09–1.62) | ||
| rs1799724 | Genotype | |||||||||
| CC | 462 (73.1) | 436 (72.1) | 368 (60.0) | 430 (71.3) | ||||||
| CT | 160 (25.3) | 154 (25.5) | 0.889 | 0.98 (0.76–1.27) | 206 (33.6) | 157 (26.0) | 0.001 | 1.53 (1.19–1.97) | ||
| TT | 10 (1.6) | 15 (2.5) | 0.257 | 0.63 (0.28–1.41) | 39 (6.4) | 16 (2.7) | 0.001 | 2.87 (1.58–5.22) | ||
| Allele | ||||||||||
| T | 1084 (85.8) | 1026 (84.8) | 942 (76.8) | 1017 (84.3) | ||||||
| C | 180 (14.2) | 184 (15.2) | 0.499 | 0.93 (0.74–1.16) | 284 (23.2) | 189 (15.7) | <0.001 | 1.62 (1.32–1.99) | ||
| rs1800629 | Genotype | |||||||||
| GG | 541 (85.6) | 510 (84.3) | 587 (95.8) | 560 (93.0) | ||||||
| GA | 88 (13.9) | 92 (15.2) | 0.526 | 0.90 (0.66–1.24) | 25 (4.1) | 42 (7.0) | 0.028 | 0.57 (0.34–0.94) | ||
| AA | 3 (0.5) | 3 (0.5) | 0.954 | 0.95 (1.91–4.75) | 1 (0.2) | 0 (0) | — | |||
| Allele | ||||||||||
| G | 1170 (92.6) | 1112 (91.9) | 1199 (97.8) | 1162 (96.5) | ||||||
| A | 94 (7.4) | 98 (8.1) | 0.545 | 0.91 (0.68–1.23) | 27 (2.2) | 42 (3.5) | 0.056 | 0.62 (0.38–1.01) | ||
| rs361525 | Genotype | |||||||||
| GG | 607 (96.0) | 582 (96.2) | 559 (91.2) | 553 (91.7) | ||||||
| GA | 24 (3.8) | 22 (3.6) | 0.892 | 1.04 (0.58–1.88) | 52 (8.5) | 48 (8.0) | 0.718 | 1.08 (0.72–1.63) | ||
| AA | 1 (0.2) | 1 (0.2) | 0.984 | 0.97 (0.06–15.64) | 2 (0.3) | 2 (0.3) | 0.989 | 0.99 (0.14–7.04) | ||
| Allele | ||||||||||
| G | 1238 (97.9) | 1186 (98.0) | 1170 (95.4) | 1154 (95.7) | ||||||
| A | 26 (2.1) | 24 (2.0) | 0.908 | 1.03 (0.59–1.81) | 56 (4.6) | 52 (4.3) | 0.740 | 1.07 (0.73–1.57) | ||
Abbreviation: SNPs, single nucleotide polymorphisms; CI, confidence interval; OR, odds ratio.
#Adjusted by age and sex status.
Table 4.
Association between genotype of cytokine genes and TB in the two populations.
| Gene/SNPs | Han population | Tibetan population | ||
|---|---|---|---|---|
| P # | OR# (95% CI) | P # | OR# (95% CI) | |
| IL1B | ||||
| rs1143634 | ||||
| Dominant model | 0.130 | 1.61 (0.87–2.97) | 0.154 | 1.40 (0.88–2.21) |
| Recessive model | — | — | ||
| rs16944 | ||||
| Dominant model | 0.042 | 0.78 (0.61–0.99) | 0.383 | 0.89 (0.69–1.56) |
| Recessive model | 0.098 | 0.79 (0.59–1.05) | 0.660 | 0.95 (0.73–1.22) |
| rs1143623 | ||||
| Dominant model | 0.414 | 1.11 (0.87–1.40) | 0.508 | 1.09 (0.84–1.41) |
| Recessive model | 0.123 | 1.26 (0.94–1.68) | 0.186 | 1.19 (0.92–1.55) |
| IL6 | ||||
| rs17147230 | ||||
| Dominant model | 0.193 | 1.20 (0.91–1.59) | 0.934 | 0.99 (0.77–1.27) |
| Recessive model | 0.725 | 1.04 (0.82–1.33) | 0.128 | 0.81 (0.62–1.06) |
| rs1800795 | ||||
| Dominant model | 0.161 | 0.32 (0.06–1.58) | 0.121 | 3.47 (0.72–16.79) |
| Recessive model | — | — | ||
| rs2069837 | ||||
| Dominant model | 0.044 | 0.78 (0.62–0.99) | 0.820 | 1.03 (0.82–1.29) |
| Recessive model | 0.462 | 0.82 (0.48–1.40) | 0.621 | 0.90 (0.60–1.36) |
| TNF | ||||
| rs1799964 | ||||
| Dominant model | 0.017 | 1.33 (1.05–1.68) | 0.004 | 1.40 (1.11–1.76) |
| Recessive model | 0.158 | 1.53 (0.85–2.74) | 0.627 | 1.12 (0.72–1.74) |
| rs1800630 | ||||
| Dominant model | 0.002 | 1.46 (1.15–1.85) | 0.017 | 1.34 (1.05–1.70) |
| Recessive model | 0.652 | 1.13 (0.67–1.88) | 0.421 | 1.30 (0.69–2.46) |
| rs1799724 | ||||
| Dominant model | 0.686 | 0.95 (0.74–1.22) | <0.001 | 1.65 (1.30–2.10) |
| Recessive model | 0.263 | 0.63 (0.28–1.41) | 0.003 | 2.49 (1.37–4.50) |
| rs1800629 | ||||
| Dominant model | 0.528 | 0.90 (0.66–1.24) | 0.038 | 0.59 (0.36–0.97) |
| Recessive model | 0.964 | 0.96 (0.19–4.80) | — | |
| rs361525 | ||||
| Dominant model | 0.899 | 1.04 (0.58–1.85) | 0.727 | 1.07 (0.72–1.61) |
| Recessive model | 0.973 | 0.95 (0.06–15.31) | 0.982 | 0.98 (0.14–6.97) |
Abbreviation: SNP, single nucleotide polymorphism; CI, confidence interval; OR, odds ratio.
#Adjusted by sex and age.
Figure 1.
Linkage disequilibrium (LD) of cytokine gene SNPs in the both Han (above) and Tibetan (below) populations. LD r2 values (ranging from 0 to 1) for all pairs of SNPs are presented as percentages. Shading from white to black indicates the extent of LD measured as r2.
Table 5.
Haplotypes of the cytokines genes and their distributions in the two cohorts.
| Gene/haplotype | Han population | Tibetan population | ||||||
|---|---|---|---|---|---|---|---|---|
| Case(%) n = 1264 | Control(%) n = 1210 | P | OR | Case(%) n = 1226 | Control(%) n = 1210 | P | OR | |
| IL1B | ||||||||
| AGC | — | 46.4 (3.8) | 16.62 (1.4) | <0.001 | 2.82 (1.60–4.96) | |||
| GAC | 75.4 (6.0) | 86.6 (7.2) | 0.251 | 0.83 (0.60–1.42) | 91.4 (7.5) | 86.6 (7.2) | 0.785 | 1.04 (0.77–1.42) |
| GAG | 551.5 (43.6) | 492.1 (40.7) | 0.097 | 1.15 (0.98–1.35) | 615.0 (50.2) | 492.1 (40.7) | <0.001 | 1.47 (1.25–1.72) |
| GGC | 601.6 (47.6) | 605.8 (50.1) | 0.285 | 0.92 (0.78–1.08) | 466.3 (38.0) | 605.8 (50.1) | <0.001 | 0.61 (0.52–0.72) |
| Other* | 35.4 (2.8) | 25.6 (2.1) | ? | ? | 7.0 (0.5) | 8.9 (0.7) | ? | ? |
| IL6 | ||||||||
| AGA | 674.3 (53.3) | 615.9 (50.9) | 0.259 | 1.01 (0.94–1.28) | 531.0 (43.3) | 615.9 (50.9) | <0.001 | 0.74 (0.63–0.87) |
| AGG | 34.73 (2.7) | 39.11 (3.2) | 0.470 | 0.84 (0.53–1.34) | 26.62 (2.2) | 39.1 (3.2) | 0.108 | 0.67 (0.40–1.10) |
| TGA | 372.8 (29.5) | 345.1 (22.7) | 0.633 | 1.04 (0.88–1.24) | 367.0 (29.9) | 345.1 (28.5) | 0.428 | 1.07 (0.90–1.28) |
| TGG | 180.3 (14.3) | 203.9 (16.9) | 0.070 | 0.82 (0.66–1.02) | 293.4 (23.9) | 203.9 (16.9) | <0.001 | 1.56 (1.27–1.90) |
| Other* | 2.0 (0.2) | 6.0 (0.5) | ? | ? | 8.0 (0.6) | 6 (0.5) | ? | ? |
| TNF | ||||||||
| CACGG | 189.0 (15.6) | 242.9 (19.2) | 0.018 | 0.78 (0.63–0.96) | 271.3 (22.1) | 189.0 (15.6) | <0.001 | 1.54 (1.25–1.89) |
| CCCGA | — | 53.3 (4.3) | 22.0 (1.8) | <0.001 | 2.46 (1.49–4.07) | |||
| TCCAG | 95.5 (7.9) | 93.9 (7.4) | 0.666 | 1.07 (0.79–1.44) | 25.2 (2.1) | 95.5 (7.9) | <0.001 | 0.25 (0.16–0.38) |
| TCCGG | 716.6 (59.2) | 718.1 (56.8) | 0.221 | 1.11 (0.94–1.30) | 591.2 (48.2) | 716.6 (59.2) | <0.001 | 0.64 (0.55–0.75) |
| TCTGG | 181.5 (0.2) | 180.0 (14.2) | 0.595 | 1.06 (0.85–1.33) | 277.8 (22.7) | 181.5 (15.0) | <0.001 | 1.67 (1.35–2.05) |
| Other* | 27.6 (2.3) | 29.1 (2.3) | 7.2 (0.5) | 5.6 (0.5) | ||||
CI, confidence interval; OR, odds ratio.
*Haplotypes with frequency <0.03 were pooled into this category.
To validate the aforementioned results, we performed an independent study in a Chinese Tibetan population (Tables 3 and 4). Among the three SNPs analyzed in IL1B, the G allele (P = 0.023) and GG genotype (P = 0.028) of rs16944 were more prevalent in controls than in TB patients. No significant difference was identified in the distributions of IL6 genotypes in TB and control groups. In TNF, the rs1799964 C allele (P = 0.011) and CT genotype (P = 0.004) as well as the rs1800630 A allele (P = 0.005) and CA genotype (P = 0.002) increased risk of TB. SNPs in TNF were also associated with TB under other genetic models: rs1799964 (dominant: P = 0.004), rs1800630 (dominant: P = 0.017), rs1799724 (dominant: P < 0.001; recessive: P = 0.003) and rs1800629 (dominant: P = 0.038). The LD between the SNPs is shown in Fig. 1. In the haplotype analysis, a total of three haplotypes in IL1B, two in IL6 and five in TNF were found to be associated with TB (Table 5).
Examination of smoking-specific effects with TB
We also determined whether genetic determinants of TB were smoking-dependent in the Chinese Han population by performing an allele-smoking interaction analysis. As shown in Table 6, TNF rs1800630 (P = 0.026) was associated with TB in the non-smoking group, but not in the smoking group. In addition, 5 SNPs (IL1B, rs16944: P < 0.001; IL6, rs1800795: P < 0.001; rs2069837: P < 0.001; TNF, rs1800629: P = 0.001; rs361525: P < 0.001) within the three genes were associated with smoking TB.
Table 6.
Association of cytokine genes with TB stratified by smoking status.
| Gene/SNPs | Genetic model | Non-smoking | Genetic model | Smoking | ||
|---|---|---|---|---|---|---|
| P # | OR# (95% CI) | P # | OR# (95% CI) | |||
| IL1B | Allele | Allele | ||||
| rs1143634G > A | 0.166 | 1.63 (0.82–3.23) | 0.986 | — | ||
| rs16944G > A | 0.883 | 1.02 (0.83–1.24) | <0.001 | 153.50 (37.79–623.61) | ||
| rs1143623C > G | 0.430 | 1.08 (0.89–1.32) | 0.451 | 1.12 (0.84–1.50) | ||
| IL6 | Allele | Allele | ||||
| rs17147230A > T | 0.565 | 0.94 (0.77–1.15) | 0.130 | 1.26 (0.94–1.69) | ||
| rs1800795G > C | 0.226 | 0.26 (0.031–2.28) | <0.001 | 0.001 (0–0.10) | ||
| rs2069837A > G | 0.046 | 0.77 (0.59–1.00) | <0.001 | 59.03 (8.18–426.17) | ||
| TNF | Allele | Allele | ||||
| rs1799964T > C | 0.017 | 1.35 (1.05–1.73) | 0.427 | 0.86 (0.60–1.24) | ||
| rs1800630C > A | 0.026 | 1.34 (1.04–1.72) | 0.653 | 0.92 (0.62–1.34) | ||
| rs1799724C > T | 0.804 | 0.97 (0.73–1.27) | 0.654 | 0.91 (0.60–1.38) | ||
| rs1800629G > A | 0.156 | 0.76 (0.52–1.11) | 0.001 | 0.48 (0.30–0.75) | ||
| rs361525G > A | 0.961 | 1.02 (0.47–2.21) | <0.001 | 0.26 (0.13–0.56) | ||
SNPs, single nucleotide polymorphisms; CI, confidence interval; OR, odds ratio.
#Adjusted by age and sex.
Discussion
Understanding the genetic factors underlying TB has attracted increasing attention. In this study, we analyzed the association between polymorphisms in three cytokine genes and TB in two independent studies. We found the IL1B rs16944 was associated with TB susceptibility in the two studies. Significant associations were also found for both rs1799964 and rs1800630 in TNF with TB, which were validated in the Tibetan population.
IL1B is a member of the IL1 cytokine family, which has an important role in the initiation and propagation of immune and inflammatory reactions. IL1B is a typical proinflammatory cytokine which can promote both local and systemic responses41. It was shown that alveolar macrophages from active TB subjects secreted high levels of IL1B42. Additionally, the production of this cytokine was associated with severity of human TB43. In addition, IL1B was reported to play an important role in the pathogenesis of TB in mice and human subjects44,45. rs1143634 and rs16944 are functional SNPs in IL1B. rs16944 has been associated with some diseases such as esophageal cancer46, inflammatory bowel disease47 and schizophrenia. However, rs16944 was variably associated with TB among different populations. One report in a small cohort from China suggested no association between the SNP and TB disease48. A similar study conducted in Kazakhstan did not show a significant association between the risk of TB and rs1694449. Awomoyi et al. reported that rs16944 both heterozygosity and homozygosity were a protective factor for TB in Gambian50, in accordance with our results in the Chinese Han population. However, in our validation cohort, we found that rs16944 G allele and GG genotype instead of AA/GA were risk factors for TB. It was reported that rs16944 regulates IL1B levels and the homozygosity TT genotype had the highest expression of IL-1β34,38. Higher IL-1β levels could also lead to the progressive immunopathology during M. TB infection, which may result in TB progression in human45. Combined with the information above, we speculate that rs16944 SNP may influence immune reaction caused by M. TB infection by increasing IL1B levels and RNA expression. Two other SNPs (rs1143634 and rs1143623) were not associated with TB in the two cohorts. This result was the same as previous studies that were conducted in various populations16,51–53.
IL6 is involved in the immune response, inflammation, coagulation, cell differentiation and tumorigenesis. High IL6 levels were related to inflammatory diseases such as rheumatoid arthritis54. In local inflammatory reactions, IL6 induces chemotaxis to mononuclear phagocytes55. When infected with M. TB, impaired bronchial epithelial cells activate mononuclear cells and lymphocytes to produce IL6, which then produces numerous antigen immune responses56. IL6 promotes IFNγ induction in early inflammatory responses and the important role of IL6 in TB susceptibility has been identified by IL6 knock-out mice24. A recent study in the Chinese Tibetan population showed an association between the rs2069837 G allele and TB57. However, there were no studies have been researched the association between rs2069837 polymorphism and its function. Our results in the Han population are consistent with He et al.; although the observation failed to validate in our Tibetan cohort. Those inconsistent results could be explained by differences in the original of the samples. Subjects in our validation cohort were derived from the Aba Tibetan Autonomous Prefecture, and their subjects were recruited from the Tibet Autonomous Region. We also demonstrated that the other two SNPs (rs17147230 and rs1800795) were not associated with TB in the Han and Tibetan populations. More studies are warranted to validate these results in the future.
The response to M. TB infection is characterized by a strong immunocyte-mediated immune response, with the induction of TNF gene expression. TNF-alpha is important in the control of M. TB infection, and latent infection subjects with blocked TNF production can rapidly reactivate58. TNF-alpha not only plays a critical role in the immune response to TB but also participates in granuloma formation59. In our initial study, we demonstrated that rs1799964 (allele C and heterozygote CT) and rs1800630 (allele A and heterozygote CA) were protective factors against TB. The same results were also detected in our validation cohort, which strongly suggested that TNF was a causal gene for TB susceptibility. However, the r2 between rs1799964 and rs1800630 was 0.87 and 0.81, respectively in the Han and Tibetan cohort, signifying that the association of TB with rs1800630 may be due to its LD with rs1799964 or vice versa. Studies have demonstrated that the rs1799964 and rs1800630 could increase the TNF RNA expression and secretion of the TNF-alpha protein31,36,39. In the present study, the same genotypes were associated with TB disease. Therefore, we speculate that these two SNPs may control the progression of TB disease by increasing the RNA expression and cytokine levels of TNF. In addition, overexpression of TNF-alpha has been implicated in the immunopathology of TB, such as tissue necrotising reactions which are important for the transmission of M. TB60,61. However, previous study proposed that other genes polymorphisms were also found to control the production of the cytokines included in our study62. Thus, a limitation of our study was that we did not perform functional validation of the significant SNPs.
A published study in the Chinese Uygur population observed that rs1799964 was associated with TB drug-resistance whereas it was not related to TB susceptibility63. They also found the rs1800630 AA + CA genotype was a risk factor for TB. The significant association of rs1799724 and rs1800629 with TB in our Tibetan cohort was the same as a previous study63–65. A published meta-analysis suggested that rs361525 was not associated with TB66 and our study in both the Han and Tibetan populations also reports the same result. Since some of our significant findings were only detected in dominant/recessive models calculated by logistic regressions, correction for multiple comparisons such as Bonferroni may be appropriate to reduce a type I error in the data analysis. However, limiting the type I error may simultaneously increase the type II error67. In addition, the primary finding in this study was the significant association between TNF polymorphisms and TB disease in allele/genotype models and the association did not change in dominant/recessive models. What’s more, this association was validated in the Tibetan population, which suggested that this finding was not due to chance. Moreover, the rs1800630 showed the same effect on TB disease in previous studies as ours (Table S2). Therefore, Bonferroni correction may have limited utility in our study.
As mentioned above, all of the three cytokine genes were associated with TB in the initial study. However, only TNF polymorphisms rs1799964 and rs1800630 were fully validated under the allele/genotype model in the Tibetan group. Thus, we speculated that TNF has a more critical role in TB risk than other two cytokines. Meanwhile, the contradictory results in our study are also worth consideration. These discrepant results may be due to the differences in genetic and environmental factors between the Han and Tibetan populations. Firstly, the MAF and genetic background between the two cohorts were different, which may have caused the different genetic association results. Secondly, many lifestyle factors were different between the two populations. Most Tibetans live in the plateau area and their staple foods are barley, beef and butter. However, the effect of the environment on our association results is unknown.
In the Tibetan cohort, the HWE test for rs1800630 was P = 0.024 in the control group. This deviation was not found in the case group. To detect if there were any genotyping errors, we repeated genotyping in 5% of the samples, and the results remained the same. The deviation from HWE could be explained by natural selection. In 1949, geneticist JBS Haldane proposed that infectious diseases have been the primary means of natural selection during the past 5000 years68.
Stratified analyses were performed on the 11 SNPs based on smoking status at TB onset in the Chinese Han population (subjects were divided into smoking and non-smoking). Interestingly, we found that the associations of IL1B and IL6 polymorphisms were more pronounced among smokers, which was similar to a previous study of TB69. Besides, SNPs within TNF revealed significant differences in allele frequencies between TB and controls in both smoking and non-smoking groups. Smoking is likely to be a risk factor for TB progression70. Our results further underline the critical role of smoking at TB onset as an important factor to consider in future TB association studies to reduce TB phenotypic heterogeneity.
In our study, a total of three haplotypes in IL1B, two in IL6 and five in TNF were strongly associated with TB in the Tibetan population. In addition, one haplotype in TNF showed significant differences between cases and control in the Han population. However, the results of haplotypes in the two cohorts were inconsistent. Since haplotype-based methods were based on the association between a polymorphism and the ancestral haplotype71, we speculate the discrepant results are likely attributable to differences in demographic history72.
Nevertheless, several limitations should be addressed in this study. First, nearly all the OR we observed were <2, which suggested that the power of the study may be inadequate to detect a role of the three gene polymorphisms on TB. However, our sample size was larger than most of the previous genetic association studies (Table S1). Second, functional validation of the meaningful SNPs was not performed. Thus the mechanism underlying the genetic association is unknown.
Conclusion
In summary, our study demonstrated that one SNP (rs16944) in IL1B and two SNPs (rs1799964 and rs1800630) in TNF were associated with susceptibility to or protection against TB development in our two studies. In addition, IL6 rs2069837 was a risk factor for TB in the Chinese population. Furthermore, TNF rs1799724 and rs1800629 were associated with TB in the Tibetan cohort. Our study could enhance the understanding of TB pathogenesis for clinicians and may improve the diagnosis of TB in the future. Further studies in other ethnic groups are needed to fully validate these results to disclose the potential function of these SNPs.
Methods
Study population
In the initial cohort, a total of 636 TB patients and 608 healthy controls were included from the West China Hospital of Sichuan University. An independent validation cohort including 613 TB patients and 603 healthy was enrolled from the Aba Tibetan Autonomous Prefecture. TB cases were diagnosed by two independent experienced respiratory physicians based on sputum smear tests, sputum culture, clinical symptom, radiological, histological pathologic examination and positive response to anti-TB therapy. Healthy controls were who had no TB-related symptoms, previous of history of TB and chest x-ray signs of disease. However, most of the participants lack the detection of Tuberculin Skin Test (TST) and Interferon Gamma Release Assay (IGRA).
Participants with HIV, hepatitis B and C, cancer and immune-related diseases were excluded from this study. All of the study subjects were unrelated. Specialized nurses drew 2–5 mL of blood from each study participant into tubes containing EDTA. Blood sample was then preserved in an −80 °C freezer for further DNA extraction and genotyping. Written informed consent was obtained from each participant. All experimental procedures were done in a BSL2 laboratory. This study was approved by the Ethics Committees of the West China Hospital of Sichuan University and the People’s Hospital of the Aba Tibetan Autonomous Prefecture. All experimental procedures were conducted in accordance with the approved guidelines and regulations and the Declaration of Helsinki.
SNP selection and genotyping
Candidate SNPs for this study were selected according to the literature review of previous studies and in silico functional prediction from the NIH FuncPred website (https://snpinfo.niehs.nih.gov/snpinfo/snpfunc.html). SNPs were selected if they were reported to be associated with disease and/or predicted to have effects on function. Genomic DNA was isolated from 250 μL of blood using a genomic DNA purification kit according to the manufacturer’s instructions (Axygen Scientific Inc, Union City, CA, USA). SNPs were genotyped using the improved multiplex ligase detection reaction (iMLDR), with technical support from the Shanghai Genesky Biotechnology Company. A subgroup of 5% of the samples was repeated by iMLDR to confirm reproducibility.
Statistical analysis
Hardy-Weinberg equilibrium (HWE) proportion tests were used to evaluate the quality of the genotype data. Continuous variables were calculated by Student t-test and categorical variables were assessed by χ2-test in our study. The distribution of genotypes/alleles between cases and controls were compared by computing the odds ratio (OR) and 95% confidence intervals (95% CI) using logistic regression analyses. ORs were defined with respect to the case groups i.e. ORs >1 represent increased risk of TB. Linkage disequilibrium (LD) measure r2 was calculated and haplotype blocks were estimated using the SHEsis online software platform (http://analysis.bio-x.cn). The power of our study design was assessed by using the Power and Sample Size Calculation software73. We calculated the power for each SNPs under an allelic model as described previously74. The frequency of each SNP in the two cohorts was sufficient to provide reasonable power (>80%) to detect an allele/genotype effect with OR 2.0 or above (Table 7). Data were considered statistically significant if P < 0.05. All analyses were performed utilizing the Statistical Package for the Social Sciences (SPSS, SPSS Inc., Chicago, IL, USA), version 19.0.
Table 7.
Power of the study with different odds ratios under the allele model.
| SNPs | Power in Han | Power in Tibetan | ||||||
|---|---|---|---|---|---|---|---|---|
| MAF | OR = 2 | OR = 3 | OR = 4 | MAF | OR = 2 | OR = 3 | OR = 4 | |
| IL1B | ||||||||
| rs1143634 | 0.02 | 0.81 | 0.99 | 1 | 0.03 | 0.92 | 1 | 1 |
| rs16944 | 0.48 | 1 | 1 | 1 | 0.42 | 1 | 1 | 1 |
| rs1143623 | 0.41 | 1 | 1 | 1 | 0.48 | 1 | 1 | 1 |
| IL6 | ||||||||
| rs17147230 | 0.44 | 1 | 1 | 1 | 0.48 | 1 | 1 | 1 |
| rs1800795 | 0.002 | 0.15 | 0.34 | 0.55 | 0.002 | 0.15 | 0.34 | 0.55 |
| rs2069837 | 0.17 | 1 | 1 | 0.26 | 1 | 1 | 1 | |
| TNF | ||||||||
| rs1799964 | 0.17 | 1 | 1 | 1 | 0.22 | 1 | 1 | 1 |
| rs1800630 | 0.16 | 1 | 1 | 1 | 0.18 | 1 | 1 | 1 |
| rs1799724 | 0.14 | 1 | 1 | 1 | 0.16 | 1 | 1 | 1 |
| rs1800629 | 0.07 | 0.99 | 1 | 1 | 0.02 | 0.80 | 0.99 | 1 |
| rs361525 | 0.02 | 0.81 | 0.99 | 1 | 0.04 | 0.97 | 1 | 1 |
SNP, single nucleotide polymorphism; TB, tuberculosis; OR, odds ratio; MAF, minor allele frequency.
Supplementary information
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 81170042 and Grant No. 81370121), and the National Scientific and Technological Major Project of China (Grant No. 2012ZX10004-901).
Author Contributions
J.Q.H. conceived and designed the experiments. S.Q.W., M.G.W. and Y.W. performed the experiments. S.Q.W., Y.W. analyzed the data. J.Q.H., S.Q.W. wrote the paper.
Data Availability
The data from this study are available online (https://figshare.com/s/33c072e600546dc1ba14).
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-019-39249-4.
References
- 1.World Health Organization, 2017. Global tuberculosis report 2017.
- 2.The fifth national tuberculosis epidemiological survey in 2010. Chinese Journal of Antituberculosis (2012).
- 3.Zumla A, Raviglione M, Hafner R, von Reyn CF. Tuberculosis. N Engl J Med. 2013;368:745–755. doi: 10.1056/NEJMra1200894. [DOI] [PubMed] [Google Scholar]
- 4.Longhi RM, Zembrzuski VM, Basta PC, Croda J. Genetic polymorphism and immune response to tuberculosis in indigenous populations: a brief review. Braz J Infect Dis. 2013;17:363–368. doi: 10.1016/j.bjid.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lawn SD, Zumla AI. Tuberculosis. Lancet. 2011;378:57–72. doi: 10.1016/S0140-6736(10)62173-3. [DOI] [PubMed] [Google Scholar]
- 6.Naranbhai, V. The Role of Host Genetics (and Genomics) in Tuberculosis. Microbiol Spectr4, 10.1128/microbiolspec.TBTB2-0011-2016 (2016). [DOI] [PubMed]
- 7.Mansouri F, Heydarzadeh R, Yousefi S. The association of interferon-gamma, interleukin-4 and interleukin-17 single-nucleotide polymorphisms with susceptibility to tuberculosis. APMIS. 2018;126:227–233. doi: 10.1111/apm.12810. [DOI] [PubMed] [Google Scholar]
- 8.Hu Y, et al. Association between cytokine gene polymorphisms and tuberculosis in a Chinese population in Shanghai: a case-control study. BMC immunology. 2015;16:8. doi: 10.1186/s12865-015-0071-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu G, et al. Association between IL12B polymorphisms and tuberculosis risk: a meta-analysis. Infect Genet Evol. 2014;21:401–407. doi: 10.1016/j.meegid.2013.12.008. [DOI] [PubMed] [Google Scholar]
- 10.Areeshi, M. Y. et al. IL-10 -1082 A > G (rs1800896) polymorphism confers susceptibility to pulmonary tuberculosis in Caucasians but not in Asians and Africans: a meta-analysis. Biosci Rep37, 10.1042/BSR20170240 (2017). [DOI] [PMC free article] [PubMed]
- 11.Zhang YF, Zhao AD. Common Polymorphisms in IL-27 Genes May Contribute to Risk of Various Human Diseases in Asian Populations: A Meta-Analysis. Med Sci Monit. 2016;22:766–775. doi: 10.12659/MSM.895558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu H, et al. The association between the CC chemokine ligand 5 -28C > G gene polymorphism and tuberculosis susceptibility. Saudi Med J. 2015;36:1400–1407. doi: 10.15537/smj.2015.12.12264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yim JJ, Selvaraj P. Genetic susceptibility in tuberculosis. Respirology (Carlton, Vic.) 2010;15:241–256. doi: 10.1111/j.1440-1843.2009.01690.x. [DOI] [PubMed] [Google Scholar]
- 14.Pociot F, Mølvig J, Wogensen L, Worsaae H, Nerup J. A Taql polymorphism in the human interleukin‐ 1β (IL‐1β) gene correlates with IL‐1β secretion in vitro. European Journal of Clinical Investigation. 1992;22:396–402. doi: 10.1111/j.1365-2362.1992.tb01480.x. [DOI] [PubMed] [Google Scholar]
- 15.Santtila S, Savinainen K, Hurme M. Presence of the IL-1RA allele 2 (IL1RN*2) is associated with enhanced IL-1beta production in vitro. Scand J Immunol. 1998;47:195–198. doi: 10.1046/j.1365-3083.1998.00300.x. [DOI] [PubMed] [Google Scholar]
- 16.Meenakshi P, et al. Association of IL-1beta + 3954 C/T and IL-10-1082 G/A cytokine gene polymorphisms with susceptibility to tuberculosis. Scand J Immunol. 2013;78:92–97. doi: 10.1111/sji.12055. [DOI] [PubMed] [Google Scholar]
- 17.Engele M, et al. Induction of TNF in human alveolar macrophages as a potential evasion mechanism of virulent Mycobacterium tuberculosis. J Immunol. 2002;168:1328–1337. doi: 10.4049/jimmunol.168.3.1328. [DOI] [PubMed] [Google Scholar]
- 18.Aguillon JC, et al. Could single-nucleotide polymorphisms (SNPs) affecting the tumour necrosis factor promoter be considered as part of rheumatoid arthritis evolution? Immunobiology. 2006;211:75–84. doi: 10.1016/j.imbio.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 19.Zhou Y, et al. Polymorphisms in the SP110 and TNF-alpha Gene and Susceptibility to Pulmonary and Spinal Tuberculosis among Southern Chinese Population. Dis Markers. 2017;2017:4590235. doi: 10.1155/2017/4590235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng, M. et al. Correlation between MBL2/CD14/TNF-alpha gene polymorphisms and susceptibility to spinal tuberculosis in Chinese population. Biosci Rep38, 10.1042/BSR20171140 (2018). [DOI] [PMC free article] [PubMed]
- 21.van Crevel R, Ottenhoff TH, van der Meer JW. Innate immunity to Mycobacterium tuberculosis. Clin Microbiol Rev. 2002;15:294–309. doi: 10.1128/CMR.15.2.294-309.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.el-Ahmady O, Mansour M, Zoeir H, Mansour O. Elevated concentrations of interleukins and leukotriene in response to Mycobacterium tuberculosis infection. Ann Clin Biochem. 1997;34(Pt 2):160–164. doi: 10.1177/000456329703400205. [DOI] [PubMed] [Google Scholar]
- 23.Nagabhushanam V, et al. Innate inhibition of adaptive immunity: Mycobacterium tuberculosis-induced IL-6 inhibits macrophage responses to IFN-γ. The Journal of Immunology. 2003;171:4750–4757. doi: 10.4049/jimmunol.171.9.4750. [DOI] [PubMed] [Google Scholar]
- 24.Ladel CH, et al. Lethal tuberculosis in interleukin-6-deficient mutant mice. Infect Immun. 1997;65:4843–4849. doi: 10.1128/iai.65.11.4843-4849.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mao X, et al. IL-1beta + 3953C/T, -511T/C and IL-6 -174C/G polymorphisms in association with tuberculosis susceptibility: A meta-analysis. Gene. 2015;573:75–83. doi: 10.1016/j.gene.2015.07.025. [DOI] [PubMed] [Google Scholar]
- 26.Chen H, et al. Single nucleotide polymorphisms in the human interleukin-1B gene affect transcription according to haplotype context. Hum Mol Genet. 2006;15:519–529. doi: 10.1093/hmg/ddi469. [DOI] [PubMed] [Google Scholar]
- 27.Zhou F, et al. Associations of potentially functional variants in IL-6, JAKs and STAT3 with gastric cancer risk in an eastern Chinese population. Oncotarget. 2016;7:28112–28123. doi: 10.18632/oncotarget.8492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Higuchi T, et al. Polymorphism of the 5′-flanking region of the human tumor necrosis factor (TNF)-alpha gene in Japanese. Tissue antigens. 1998;51:605–612. doi: 10.1111/j.1399-0039.1998.tb03002.x. [DOI] [PubMed] [Google Scholar]
- 29.Sharma S, Ghosh B, Sharma S. Association of TNF polymorphisms with sarcoidosis, its prognosis and tumour necrosis factor (TNF)‐α levels in Asian Indians. Clinical & Experimental Immunology. 2008;151:251–259. doi: 10.1111/j.1365-2249.2007.03564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bennet A, et al. Association of TNF-α serum levels and TNFA promoter polymorphisms with risk of myocardial infarction. Atherosclerosis. 2006;187:408–414. doi: 10.1016/j.atherosclerosis.2005.09.022. [DOI] [PubMed] [Google Scholar]
- 31.Piosik ZM, Goegebeur Y, Klitkou L, Steffensen R, Christiansen OB. Plasma TNF-alpha levels are higher in early pregnancy in patients with secondary compared with primary recurrent miscarriage. Am J Reprod Immunol. 2013;70:347–358. doi: 10.1111/aji.12135. [DOI] [PubMed] [Google Scholar]
- 32.Cui G, et al. Polymorphism of tumor necrosis factor alpha (TNF-alpha) gene promoter, circulating TNF-alpha level, and cardiovascular risk factor for ischemic stroke. J Neuroinflammation. 2012;9:235. doi: 10.1186/1742-2094-9-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Camargo JF, Correa PA, Castiblanco J, Anaya JM. Interleukin-1beta polymorphisms in Colombian patients with autoimmune rheumatic diseases. Genes Immun. 2004;5:609–614. doi: 10.1038/sj.gene.6364133. [DOI] [PubMed] [Google Scholar]
- 34.Pociot F, Molvig J, Wogensen L, Worsaae H, Nerup J. A TaqI polymorphism in the human interleukin-1 beta (IL-1 beta) gene correlates with IL-1 beta secretion in vitro. Eur J Clin Invest. 1992;22:396–402. doi: 10.1111/j.1365-2362.1992.tb01480.x. [DOI] [PubMed] [Google Scholar]
- 35.Fishman D, et al. The effect of novel polymorphisms in the interleukin-6 (IL-6) gene on IL-6 transcription and plasma IL-6 levels, and an association with systemic-onset juvenile chronic arthritis. J Clin Invest. 1998;102:1369–1376. doi: 10.1172/JCI2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodríguez-Reyes SC, et al. Increased expression of TNFA gene in Mexican patients with acute myocardial infarction and its relationship with-857 C> T and-863 C> A polymorphisms. International journal of clinical and experimental medicine. 2016;9:17596–17603. [Google Scholar]
- 37.Mao ZR, Zhang SL, Feng B. Association of IL-10 (-819T/C, -592A/C and -1082A/G) and IL-6 -174G/C gene polymorphism and the risk of pneumonia-induced sepsis. Biomarkers. 2017;22:106–112. doi: 10.1080/1354750X.2016.1210677. [DOI] [PubMed] [Google Scholar]
- 38.Hameed I, et al. Genetic variations in key inflammatory cytokines exacerbates the risk of diabetic nephropathy by influencing the gene expression. Gene. 2018;661:51–59. doi: 10.1016/j.gene.2018.03.095. [DOI] [PubMed] [Google Scholar]
- 39.Nourian M, et al. Evaluation of tumor necrosis factor (TNF)-alpha mRNA expression level and the rs1799964 polymorphism of the TNF-alpha gene in peripheral mononuclear cells of patients with inflammatory bowel diseases. Biomed Rep. 2017;6:698–702. doi: 10.3892/br.2017.908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pappachan JV, et al. Mortality in adult intensive care patients with severe systemic inflammatory response syndromes is strongly associated with the hypo-immune TNF -238A polymorphism. Immunogenetics. 2009;61:657–662. doi: 10.1007/s00251-009-0395-6. [DOI] [PubMed] [Google Scholar]
- 41.Gabay C, Lamacchia C, Palmer G. IL-1 pathways in inflammation and human diseases. Nat Rev Rheumatol. 2010;6:232–241. doi: 10.1038/nrrheum.2010.4. [DOI] [PubMed] [Google Scholar]
- 42.Wang CH, Kuo HP. Nitric oxide modulates interleukin-1beta and tumour necrosis factor-alpha synthesis, and disease regression by alveolar macrophages in pulmonary tuberculosis. Respirology (Carlton, Vic.) 2001;6:79–84. doi: 10.1046/j.1440-1843.2001.00302.x. [DOI] [PubMed] [Google Scholar]
- 43.Chensue SW, Davey MP, Remick DG, Kunkel SL. Release of interleukin-1 by peripheral blood mononuclear cells in patients with tuberculosis and active inflammation. Infect Immun. 1986;52:341–343. doi: 10.1128/iai.52.1.341-343.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mishra BB, et al. Nitric oxide controls the immunopathology of tuberculosis by inhibiting NLRP3 inflammasome-dependent processing of IL-1beta. Nat Immunol. 2013;14:52–60. doi: 10.1038/ni.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang G, et al. Allele-specific induction of IL-1beta expression by C/EBPbeta and PU.1 contributes to increased tuberculosis susceptibility. PLoS pathogens. 2014;10:e1004426. doi: 10.1371/journal.ppat.1004426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng L, et al. Interleukin 1B rs16944 G> A polymorphism was associated with a decreased risk of esophageal cancer in a Chinese population. Clin Biochem. 2013;46:1469–1473. doi: 10.1016/j.clinbiochem.2013.05.050. [DOI] [PubMed] [Google Scholar]
- 47.Nemetz A, et al. IL1B gene polymorphisms influence the course and severity of inflammatory bowel disease. Immunogenetics. 1999;49:527–531. doi: 10.1007/s002510050530. [DOI] [PubMed] [Google Scholar]
- 48.Sun H, et al. A method of oligochip for single nucleotide polymorphism genotyping in the promoter region of the interleukin-1 beta gene and its clinical application. Oligonucleotides. 2007;17:336–344. doi: 10.1089/oli.2007.0071. [DOI] [PubMed] [Google Scholar]
- 49.Yerezhepov, D. et al. Genetic Diversity of IFγ, IL1β, TLR2, and TLR8 Loci in Pulmonary Tuberculosis in Kazakhstan. Central Asian Journal of Global Health3 (2014). [DOI] [PMC free article] [PubMed]
- 50.Awomoyi AA, et al. Polymorphism in IL1B: IL1B-511 association with tuberculosis and decreased lipopolysaccharide-induced IL-1beta in IFN-gamma primed ex-vivo whole blood assay. Journal of endotoxin research. 2005;11:281–286. doi: 10.1179/096805105X58706. [DOI] [PubMed] [Google Scholar]
- 51.Kusuhara K, Yamamoto K, Okada K, Mizuno Y, Hara T. Association of IL12RB1 polymorphisms with susceptibility to and severity of tuberculosis in Japanese: a gene-based association analysis of 21 candidate genes. International journal of immunogenetics. 2007;34:35–44. doi: 10.1111/j.1744-313X.2007.00653.x. [DOI] [PubMed] [Google Scholar]
- 52.Serezani H. Too Much of a Good Thing: Finding an IL1B Polymorphism That Increases Tuberculosis Susceptibility. Science Translational Medicine. 2014;6:260ec187–260ec187. doi: 10.1126/scitranslmed.aaa1237. [DOI] [Google Scholar]
- 53.Souza de Lima D, Ogusku MM, Sadahiro A, Pontillo A. Inflammasome genetics contributes to the development and control of active pulmonary tuberculosis. Infect Genet Evol. 2016;41:240–244. doi: 10.1016/j.meegid.2016.04.015. [DOI] [PubMed] [Google Scholar]
- 54.Wei ST, Sun YH, Zong SH, Xiang YB. Serum Levels of IL-6 and TNF-alpha May Correlate with Activity and Severity of Rheumatoid Arthritis. Med Sci Monit. 2015;21:4030–4038. doi: 10.12659/MSM.895116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Romano M, et al. Role of IL-6 and its soluble receptor in induction of chemokines and leukocyte recruitment. Immunity. 1997;6:315–325. doi: 10.1016/S1074-7613(00)80334-9. [DOI] [PubMed] [Google Scholar]
- 56.Song Q, Li H, Shao H, Li C, Lu X. MicroRNA-365 in macrophages regulates Mycobacterium tuberculosis-induced active pulmonary tuberculosis via interleukin-6. International journal of clinical and experimental medicine. 2015;8:15458–15465. [PMC free article] [PubMed] [Google Scholar]
- 57.He S, et al. Association of IL4, IL6, and IL10 polymorphisms with pulmonary tuberculosis in a Tibetan Chinese population. Oncotarget. 2018;9:16418–16426. doi: 10.18632/oncotarget.23995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Abebe M, et al. Expression of apoptosis-related genes in an Ethiopian cohort study correlates with tuberculosis clinical status. Eur J Immunol. 2010;40:291–301. doi: 10.1002/eji.200939856. [DOI] [PubMed] [Google Scholar]
- 59.Stein CM, et al. Evidence for a major gene influence on tumor necrosis factor-alpha expression in tuberculosis: path and segregation analysis. Human heredity. 2005;60:109–118. doi: 10.1159/000088913. [DOI] [PubMed] [Google Scholar]
- 60.Gardam MA, et al. Anti-tumour necrosis factor agents and tuberculosis risk: mechanisms of action and clinical management. Lancet Infect Dis. 2003;3:148–155. doi: 10.1016/S1473-3099(03)00545-0. [DOI] [PubMed] [Google Scholar]
- 61.Belay M, et al. Lipoarabinomannan-specific TNF-alpha and IFN-gamma as markers of protective immunity against tuberculosis: a cohort study in an endemic setting. APMIS. 2015;123:851–857. doi: 10.1111/apm.12423. [DOI] [PubMed] [Google Scholar]
- 62.Lachmandas E, et al. Tissue Metabolic Changes Drive Cytokine Responses to Mycobacterium tuberculosis. J Infect Dis. 2018;218:165–170. doi: 10.1093/infdis/jiy173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li Q, et al. The single nucleotide polymorphisms in TNF-α promoter are associated with susceptibility and clinical features of pulmonary tuberculosis in Chinese Uygurs. International journal of clinical and experimental medicine. 2017;10:11596–11605. [Google Scholar]
- 64.Wang Q, Zhan P, Qiu LX, Qian Q, Yu LK. TNF-308 gene polymorphism and tuberculosis susceptibility: a meta-analysis involving 18 studies. Molecular biology reports. 2012;39:3393–3400. doi: 10.1007/s11033-011-1110-x. [DOI] [PubMed] [Google Scholar]
- 65.Ma MJ, et al. Toll-like receptors, tumor necrosis factor-alpha, and interleukin-10 gene polymorphisms in risk of pulmonary tuberculosis and disease severity. Hum Immunol. 2010;71:1005–1010. doi: 10.1016/j.humimm.2010.07.009. [DOI] [PubMed] [Google Scholar]
- 66.Lee YH, Song GG. Associations between tumor necrosis factor-alpha polymorphisms and susceptibility to pulmonary tuberculosis: meta-analysis. Genetics and molecular research: GMR. 2015;14:8602–8612. doi: 10.4238/2015.July.31.8. [DOI] [PubMed] [Google Scholar]
- 67.Streiner DL, Norman GR. Correction for multiple testing: is there a resolution? Chest. 2011;140:16–18. doi: 10.1378/chest.11-0523. [DOI] [PubMed] [Google Scholar]
- 68.Lederberg J. JBS Haldane (1949) on infectious disease and evolution. Genetics. 1999;153:1–3. doi: 10.1093/genetics/153.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pan H, Dai Y, Tang S, Wang J. Polymorphisms of NOD2 and the risk of tuberculosis: a validation study in the Chinese population. International journal of immunogenetics. 2012;39:233–240. doi: 10.1111/j.1744-313X.2011.01079.x. [DOI] [PubMed] [Google Scholar]
- 70.Dye C, Williams BG. The population dynamics and control of tuberculosis. Science. 2010;328:856–861. doi: 10.1126/science.1185449. [DOI] [PubMed] [Google Scholar]
- 71.Gabriel SB, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 72.Reich DE, et al. Linkage disequilibrium in the human genome. Nature. 2001;411:199–204. doi: 10.1038/35075590. [DOI] [PubMed] [Google Scholar]
- 73.Dupont WD, Plummer WD., Jr. Power and sample size calculations for studies involving linear regression. Control Clin Trials. 1998;19:589–601. doi: 10.1016/S0197-2456(98)00037-3. [DOI] [PubMed] [Google Scholar]
- 74.Wu S, et al. Evaluation of TLR2, TLR4, and TOLLIP polymorphisms for their role in tuberculosis susceptibility. APMIS. 2018;126:501–508. doi: 10.1111/apm.12855. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data from this study are available online (https://figshare.com/s/33c072e600546dc1ba14).