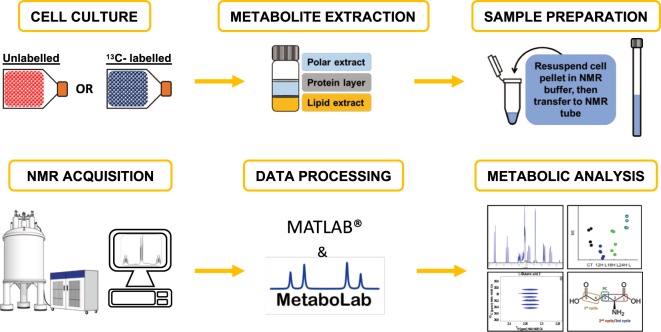
Figure 1.
Workflow for NMR tracer-based metabolism. Cell cultures are grown in a medium containing 13C or 15N- labelled precursors. Typically, 10–20 million cells are required. Adherent cells are removed from the flask surface using a scraper, thereby avoiding the use of trypsin which generates unwanted metabolites. Cells in suspension (e.g. hematological cancer cells) are harvested by centrifugation (See Materials and Methods for detailed protocols). Metabolites are obtained using a methanol/chloroform/water system where the polar extract in the upper phase is easily collected. Samples are vacuum dried and re-suspended in NMR buffer before being transferred to an NMR tube. Different NMR spectra are acquired, typically including a 1D-1H-NOESY and a 2D-1H,13C-HSQC, often also 2D-1H,1H-TOCSY spectra. For a complete isotopomer analysis, a combination of NMR and GC-MS may be used, which can be analyzed using the MetaboLab software24. From this analysis site specific label incorporations can be derived19. The spectrometer in this Figure is taken from WikiCommons, CC BY-SA 3.0 (https://creativecommons.org/licenses/by-sa/3.0/) https://commons.m.wikimedia.org/wiki/File:Nuclear_magnetic_resonance_(NMR).png.