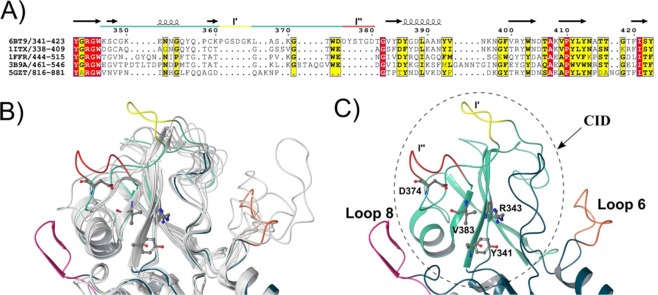
Figure 2.
Structure-based sequence alignment of ChiA74 with other chitinases. The image shows the structure-based sequence alignment of the catalytic cleft of ChiA74 and others CID containing chitinases. Similar and conserved residues are indicated in yellow and red, respectively, according with ENDScript server35. UCSF Chimera34 for structures superimposing. (A) Structure-based sequence alignment, only the region of the CID is shown. Main difference with other CIDs is the CID-loop 1 long. Regions l′ and 1″ are indicated in yellow and red, respectively. The PDB ID are the following: 6BT9 (chitinase ChiA74 from B. thuringiensis); 1ITX (catalytic domain of chitinase A1 from B. circulans WL-12); 1FFR (chitinase of mutant Y390F complexed with hexa-N-acetylchitohexaose from S. marcescens); 3B9A (V. harveyi chitinase A complexed with hexasaccharide) and 5GZT (chitinase ChiW from Paenibacillus sp. str. FPU-7). (B) Structural superimposition of ChiA74 with other chitinases where the differences between structures are shown with color. PDB codes of the superimposed structures are the same used in A. (C) CID in ChiA74 (dashed line) protrudes from Loops 6 and 8 of the TIM-barrel. Residues that would be interacting with substrate are indicated. Region 1′ of the CID-loop1 closes the substrate binding groove ″ of the active cleft (yellow).