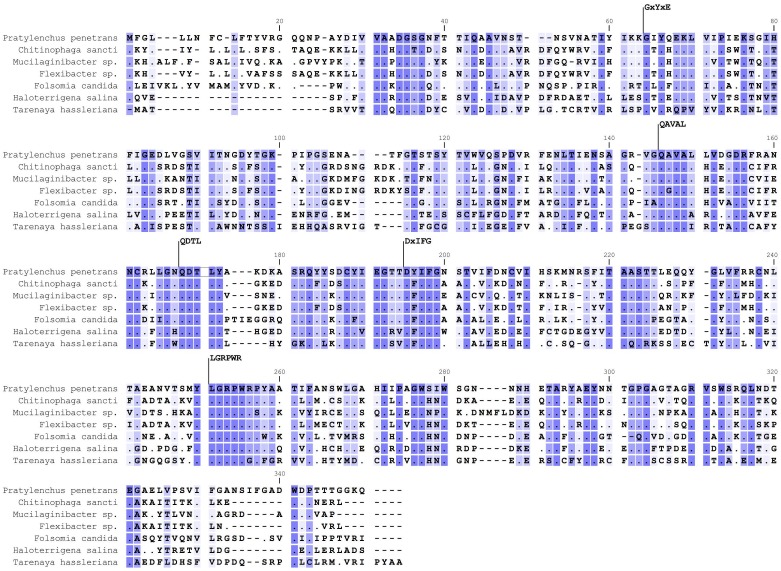
Fig 2. Multiple sequence alignment of the predicted Pp-PME protein of Pratylenchus penetrans with PMEs of other organisms.
Representative species of bacteria (Chitinophaga sancti, Mucilaginibacter sp., Flexibacter sp.), collembolan (Folsomia candida), Archaea (Haloterrigena salina) and plants (Tarenaya hassleriana) were used. The five conserved sequence segments typical of PME proteins are numbered according to their positions in the corresponding Pp-PME predicted protein (64_GxYxE, 146_QAVAL, 168_QDTL, 195_DxIFG, and 251_LGRPW). Conserved residues among species are indicated by dark blue shading and dots, whereas similar residues are represented in light blue using a threshold for shading of 50% similarity. The accession numbers corresponding to each species are presented in S3 Table.