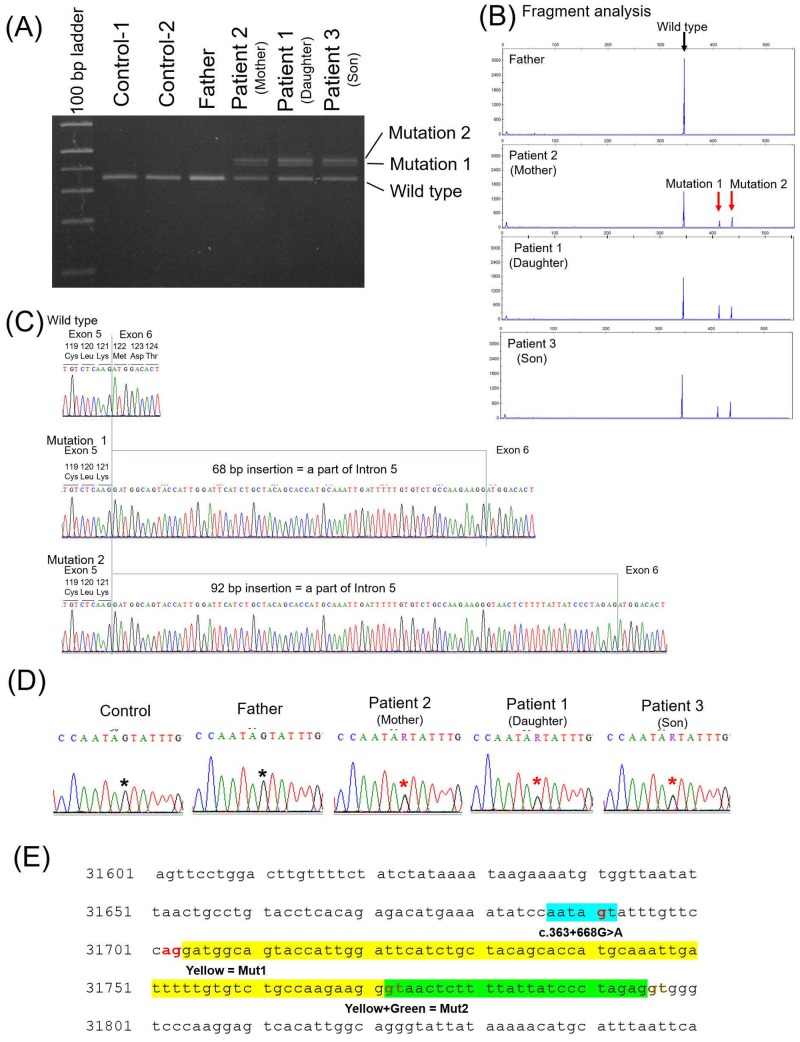
Fig 4. Analysis of RNA and introns of TSC1 and TSC2.
Three patients (patients 1, 2, and 3), their healthy father (husband of patient 2), and two healthy controls were assessed. Total RNA was extracted from peripheral blood lymphocytes and template DNA was synthesised. (A, B) Reverse transcription-PCR (RT-PCR) with the template DNA and primer pair covering exons 5–7 of TSC1. Full-length agarose gel electrophoresis of RT-PCR products is shown in S1 Fig. In lanes for all three patients, two additional, longer amplification product bands were observed (mutations 1 and 2). (C) Direct DNA sequencing with products amplified using RT-PCR revealed an insertion of 68 base pairs (bp) (mutation 1) or 92 bp (mutation 2) of a part of the intron 5 sequence between exons 5 and 6. (D) The sequences of a part of intron 5 for the three patients and their father were detected via direct sequencing of DNA extracted from peripheral blood. The three patients harboured point mutations (c.363+668G>A) in the heterojunction. (E) A section of intron 5. Intron 5 consists of 2096 bp in total. A segment of the sequence of intron 5 of TSC1 including the point mutation was detected by direct DNA sequencing. Sixty-eight base pairs (yellow) and 92 bp (yellow and green) were inserted between exons 5 and 6, as shown in (C). The 6-bp sequence including the point mutation (light blue) may serve as a new branch point. AG (red) immediately upstream of the inserted sequence may function as the splice acceptor site, and 2 GT (brown) after the inserted sequence may function as the splice donor sites. Therefore, the point mutation may cause splicing anomalies. Because 68 and 92 are not multiples of 3, the splicing anomaly may cause a frame shift and loss of function of TSC1 in the three patients.