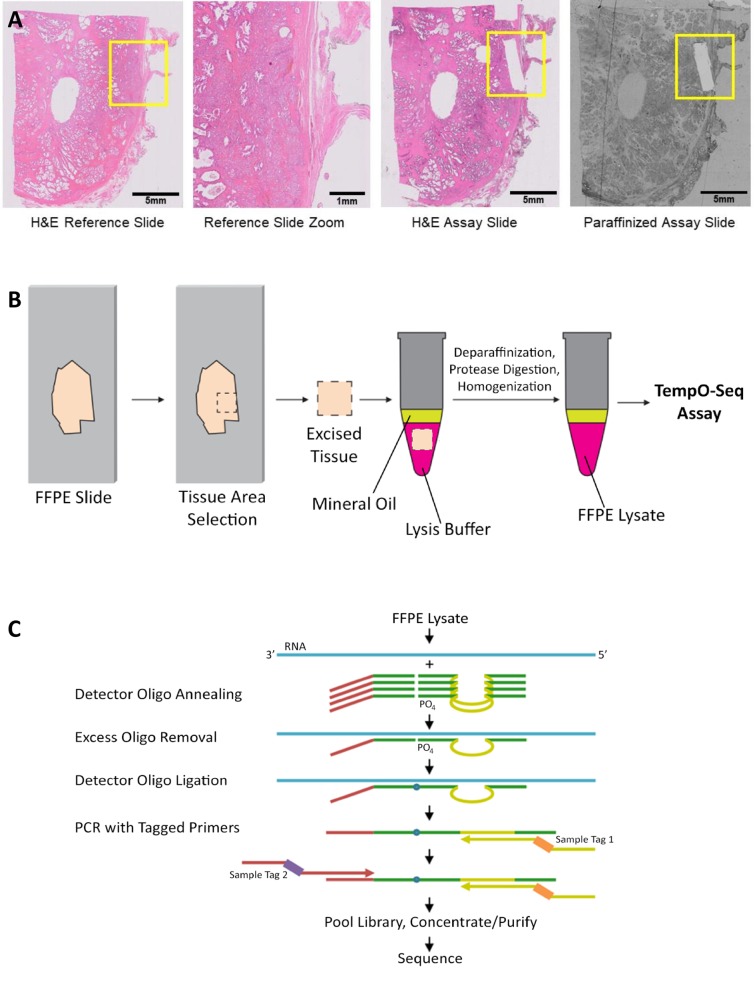
Fig 1. Processing FFPE samples for the TempO-Seq assay.
(A) Examples of slides processed through the TempO-Seq FFPE assay. Left panel shows an H&E stained section, with the yellow box indicating the area of interest. Center left shows an expanded image of this area, demonstrating the mixed histology that would affect the data if the entire area were to be scraped and profiled. Areas identified from a stained tissue section can be scraped from an unstained, paraffinized adjacent section (right), or (if RNase-free reagents are used for staining) directly from a stained section (center right). The scraped areas in this case were ~1 x 5 mm, aligned with the focal histology of interest, and sufficient for gene expression profiling. (B) An area of interest is manually scraped from mounted FFPE sections. The tissue is added directly into 1X FFPE lysis buffer, overlaid with mineral oil, and then heated at 95°C for 5 minutes. FFPE Protease is added and the sample is incubated and manually homogenized. The processed lysate is then ready for input directly into the annealing step of the TempO-Seq assay. (C) Schematic of the TempO-Seq detector oligo annealing and ligation process.