FIG 5.
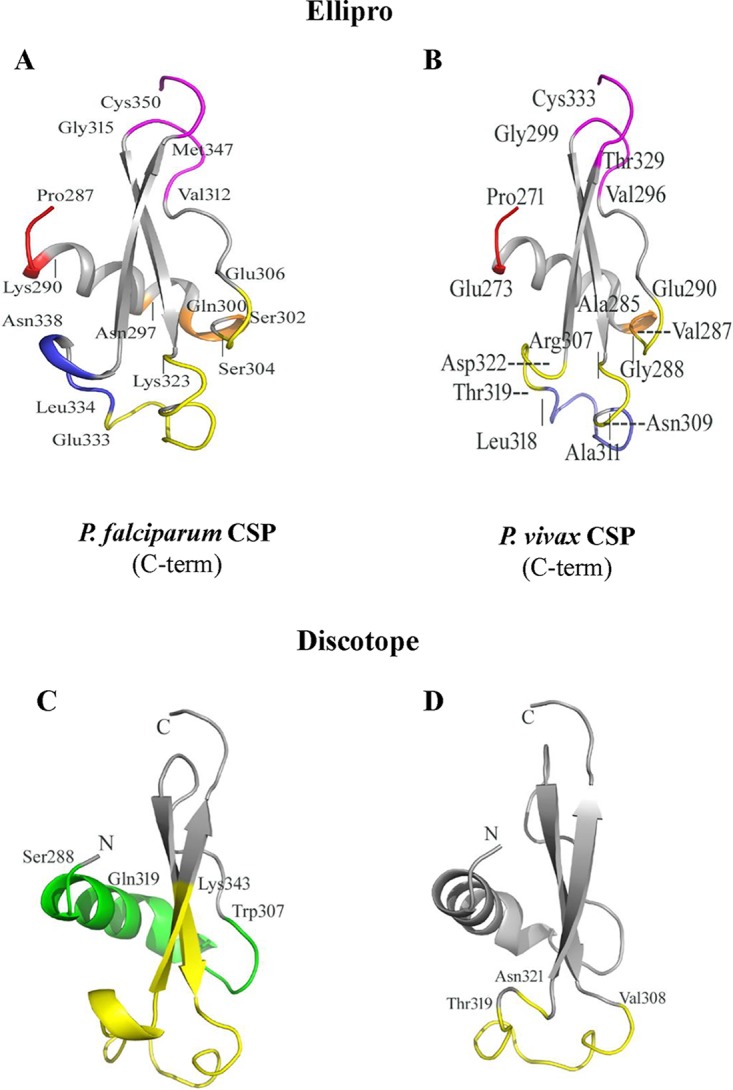
Discontinuous epitopes are predicted in similar regions on the C-terminal domains of P. vivax and P. falciparum CSPs using two separate prediction tools. (A and B) Cartoon diagrams showing the locations of conformational epitopes on the 3D structures of the C-terminal domains of P. falciparum CSP (A) and P. vivax CSP (B) as predicted by ElliPro (31). (A) ElliPro-predicted scores, in descending order, for the five PfCSP epitopes are 0.717 (magenta), 0.699 (red), 0.668 (yellow), 0.668 (orange), and 0.547 (blue). (B) The scores for the five epitopes, ranked in order, for PvCSP are 0.725 (red), 0.721 (magenta), 0.647 (slate), 0.556 (yellow), and 0.503 (orange). Individual residues are marked and described in the text. (C and D) Cartoon diagrams of conformational epitopes predicted by DiscoTope (30) for the C-terminal domains of P. falciparum and P. vivax CSPs, respectively. (C) The two predicted epitopes for PfCSP are highlighted in green for a 19-residue epitope and in yellow for a 24-residue epitope. (D) The single 13-residue predicted epitope for PvCSP is highlighted in yellow.
