A supported lipid bilayer is used as a chemical circuit board to carry out molecular computation with a network of nanoparticles.
Abstract
Using nanoparticles as substrates for computation enables algorithmic and autonomous controls of their unique and beneficial properties. However, scalable architecture for nanoparticle-based computing systems is lacking. Here, we report a platform for constructing nanoparticle logic gates and circuits at the single-particle level on a supported lipid bilayer. Our “lipid nanotablet” platform, inspired by cellular membranes that are exploited to compartmentalize and control signaling networks, uses a lipid bilayer as a chemical circuit board and nanoparticles as computational units. On a lipid nanotablet, a single-nanoparticle logic gate senses molecules in solution as inputs and triggers particle assembly or disassembly as an output. We demonstrate a set of Boolean logic operations, fan-in/fan-out of logic gates, and a combinational logic circuit such as a multiplexer. We envisage that our approach to modularly implement nanoparticle circuits on a lipid bilayer will create new paradigms and opportunities in molecular computing, nanoparticle circuits, and systems nanoscience.
INTRODUCTION
Across many length scales, matter has merged with computation, from micro-sized droplets (1) and particles (2–4) to biomolecules (5–14) and molecular machines (15). However, implementing computation in nanoparticles remains unexplored despite a wide range of applications that would benefit from algorithmically controlling their potentially useful photonic, electrical, magnetic, catalytic, and material properties that are not accessible from molecular systems (16–23). Systems of nanoparticles equipped with computing capability—nanoparticle “circuits”—can autonomously perform complex tasks in response to external stimuli, directing the flow of matter and information at the nanoscale. To date, a common approach to using nanoparticles as substrates for computation is functionalizing the particles with stimuli-responsive ligands (24–30). A group of surface-modified nanoparticles then can carry out elementary logic operations, responding to various chemical and physical inputs. Ideally, one should be able to use individual nanoparticles as modular nano-parts and implement a desired computation in a plug-and-play manner. However, the current approach has been limited to installing few logic operations that control only simple outputs, such as aggregation of particles in solution. This limitation is not because of the lack of sophistication in nanoparticles or surface ligands but because of the difficulties in modular wiring of multiple logic gates in the solution phase, where inputs, logic gates, and outputs all diffuse uncontrollably in the three-dimensional (3D) space. Specifically, the following constraints have imposed limitations on nanoparticle computing. First, particles with information-processing functions are irreversibly altered after one operation and mixed with unreacted particles or inputs in a bulk solution. The lack of compartmentalization prevents the implementation of more than one computational task per test tube. Second, it is difficult to control or analyze structural changes, dynamic interactions, and output signals of freely diffusing particles in solution. In most cases, only an ensemble-averaged signal such as a color change of solution is obtained as a final readout, which averages out particle-by-particle responses of the computation. To construct complex yet reliable nanoparticle circuits at the level of single particles, it is necessary to go beyond solution-phase approaches and make a transition into scalable, modular platforms with in situ readout and controllability, in which a circuit of interest can be systematically constructed on the basis of digital design principles.
In nature, the cell membrane is a biological equivalent of a circuit board. Hosting a variety of receptor proteins as computational units, the membrane compartmentalizes the proteins from information-rich extracellular fluids; on the fluidic membrane surface, the receptors laterally interact with each other to carry out complex functions as a network. Each receptor, as an active constituent of the biological circuit, takes chemical and physical cues as inputs such as binding events with its ligands or changes in membrane voltage and generates outputs such as conformational changes or dimerization/dissociation reactions; in addition, the membrane allows many different computing processes to occur in parallel (31–33).
Taking inspiration from cellular membranes, we demonstrate a lipid bilayer–based nanoparticle computing platform termed lipid nanotablet (LNT) that enables nanoparticles tethered to a supported lipid bilayer (SLB) to perform logic computation in a modular manner. To implement the computation, we use SLBs, which have been widely used as synthetic mimics for cell surfaces (34), as “chemical circuit boards” and program the ways tethered nanoparticles interact with one another using surface ligands. As a proof of concept, we use light-scattering plasmonic nanoparticles as circuit components, DNA as surface ligands and molecular inputs, and biotin-streptavidin interaction for tethering. Tethering of nanoparticles to a lipid bilayer provides the following features: First, nanoparticles are compartmentalized from a solution containing molecular inputs. Second, particle-to-particle interactions are confined to occur only through lateral diffusion at the fluidic 2D reaction space. Third, the laterally confined nanoparticles are tracked and analyzed in situ with single-particle resolution because a large number of light-scattering nanoparticles are confined in the focal plane of dark-field microscopy (DFM). We exploit these unique features to realize an unconventional way to carry out computation with single nanoparticles. This “nano-bio” computation, which occurs at the interface of nanostructures and biomolecules, translates molecular information in solution (input) into a dynamic assembly/disassembly of nanoparticles on a lipid bilayer (output).
RESULTS
A lipid bilayer as a circuit board for nanoparticle-based logic computation
A key component of an LNT is a flow chamber, of which the bottom substrate is coated with a lipid bilayer. Nanoparticles, whose optical signals, mobility, and surface DNA ligands are readily controllable, are biochemically tethered to the lipid bilayer surface via biotin-streptavidin interaction and monitored by DFM (Fig. 1A and fig. S1). The tethering of nanoparticles enables facile exchanges of input solutions through the flow chamber without washing out the computing elements on the SLB surface. We use three types of core particles with distinct light-scattering spectra: gold nanorods with silver shells, gold nanospheres, and silver nanospheres on gold seeds that exhibited red, green, and blue scattering signals, respectively (fig. S2). These particles are referred to as red, green, and blue nanoparticles. We prepared two classes of nanoparticles—receptor (R) and floater (F)—that differ in diffusion coefficients. Receptors are immobile on SLBs because their lateral diffusion is limited by a large number of biotinylated DNA linkers that strongly interact with streptavidins on an SLB. The biotinylated linkers functionalized 34 to 50% of the receptor surface valency. Floaters, whose linker density was 0.5%, were highly mobile on SLBs with a diffusion coefficient of ~1.0 μm2/s (fig. S3). As a result of the mobility, floaters actively interact with receptors across space and time, functioning as active units of computation. In this study, we used six types of nanoparticle circuit elements with distinct optical and kinetic states: red nano-receptors (R-NRs), red nano-floaters (R-NFs), green nano-receptors (G-NRs), green nano-floaters (G-NFs), blue nano-receptors (B-NRs), and blue nano-floaters (B-NFs). The surface DNA ligands mediate a receptor–floater interaction, taking DNA molecules as inputs and inducing the binding or unbinding of the receptor–floater complex as an output, only when the molecular inputs meet a Boolean logic condition (Fig. 1B).
Fig. 1. Single-nanoparticle logic computation on LNTs.
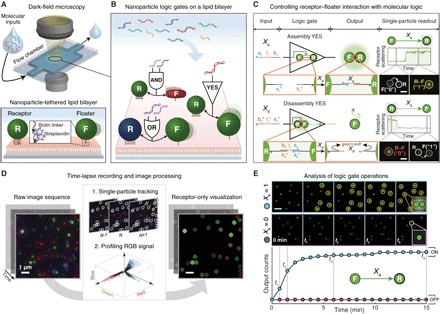
(A) Schematics of the LNT platform. Two types of DNA-modified nanoparticles, immobile receptor (R) and mobile floater (F), are tethered to an SLB and monitored by DFM. (B) R–F pairs as nanoparticle Boolean logic gates. Each logic gate takes DNA as inputs and yields either an assembly or a disassembly between the two particles as an output. Bidirectional arrows denote R–F interactions. Surface DNA ligands are not depicted. (C) Single-nanoparticle YES gates. An R–F pair acts as an Assembly YES gate when the floater binds to the receptor in response to an input DNA Xa. An R–F dimer is a Disassembly YES gate when an input Xd promotes a disassembly reaction of the dimer via toehold-mediated strand displacement. The assembly/disassembly reactions, represented by directed graph, result in step-function–like changes in receptor signals as optical readout. Functional domains are represented by color and subscripted numbers with arrowheads indicating their 3′ ends. Asterisks denote complementarity. Glowing circles behind R–F dimers illustrate plasmonic coupling. Scale bars, 1 μm. (D) Image analysis. A single-particle tracking algorithm first identifies receptor signals from a raw image sequence. Afterward, the detected signals are sampled and used to generate a new dark-field movie that visualizes only receptor signals. (E) Kinetics analysis. Receptor-only snapshots (top) and a kinetics plot (bottom) of the Assembly YES gate are provided for each input condition. A kinetics plot is obtained by cumulatively counting the number of state-switching receptors over time. Scale bar, 4 μm.
Actions of a single floater are intrinsically binary. For a given observation period, either a floater discretely switches its state (through assembly or disassembly) or it does not. Thus, controlling the “digital” actions of each floater with Boolean logic allows a receptor–floater pair to be treated as a logic gate. For the inputs of the single-nanoparticle logic gate, the logic values “0” and “1” represent the absence and presence of an input DNA in solution, respectively. For outputs, logic values are assigned to floaters and indicate whether a floater changes its state: “0” indicates a floater in its initial state, whereas “1” represents a floater whose state is switched as a result of an assembly or a disassembly reaction. Single-nanoparticle YES gates are of the simplest examples (Fig. 1C). In the Assembly YES gate, a G-NF switches its state from a diffusible monomer (“0”) to an immobile dimer (“1”) through association with a G-NR in response to a single-stranded DNA input Xa that can hybridize with surface DNA ligands of the two particles. In the Disassembly YES gate, a G-NF is initially bound to a G-NR via hybridization with an oligonucleotide Xd*. The process of forming receptor–floater dimers for Disassembly gates is termed predimerization. When a DNA input Xd is introduced, it first binds to the partially hybridized Xd* through a single-stranded toehold domain (t2) and removes the preformed DNA “bond” (Xd*) from the receptor–floater interface by fully hybridizing it. As a result, the G-NF is released from the G-NR, switching its state from an immobile dimer (“0”) to a diffusible monomer (“1”). We used this process, known as toehold-mediated strand displacement (35), to design nanoparticle Disassembly logic gates, because this simple mechanism enables robust control of DNA-modified nanoparticles using molecular interactions alone (see fig. S4 for further details). Disassembly reactions require input molecules to penetrate receptor–floater interfaces densely packed with surface DNA ligands for interactions with toehold domains, whereas assembly reactions are simply driven by collisions between receptors and floaters that are modified with input strands. Assembly reactions are thus kinetically more favorable than disassembly reactions. We use relatively high input concentration for disassembly reactions to compensate the difference in kinetics. To represent behaviors of logic-gated nanoparticles in a simple diagram, we use “nanoparticle reaction graph” abstraction. This abstraction is based on a directed graph where a node is represented by a nanoparticle and an edge is represented by logic, inputs, and reaction type. We depict an assembly reaction by a solid arrow directed from a floater to a receptor and a disassembly reaction by a dashed arrow from a receptor to a floater, as shown in Fig. 1C. The use of directed graph provides an intuitive view on how each nanoparticle logic gate behaves.
To analyze the state-switching behaviors of floaters, we monitor the signal changes of receptors, whose stationary positions allow facile monitoring of assembly and disassembly events. On the LNT platform, receptors are used as optical reporters of computation that provide single-particle–level, digital signal readout. For example, the Assembly YES gate produces output “1” when a G-NF assembles onto a G-NR through Xa. As shown in the time trace of a receptor-scattering signal and its associated DFM images in Fig. 1C, the assembly process results in a plasmonic coupling–induced, step-function–like increase in green intensity of the G-NR. The real-time operations of two YES gates are described in movie S1 (Assembly YES) and movie S2 (Disassembly YES). Scattering signals of a nanoparticle logic gate depend on plasmonic coupling between two core particles that compose the gate (36); for a given assembly or disassembly reaction, there are nine different patterns of scattering signal changes based on combinatorial plasmonic couplings induced by three different floaters (R-NF, G-NF, or B-NF) binding to three distinct receptors (R-NR, G-NR, or B-NR). Thus, multiple nanoparticle logic gates can be implemented and analyzed in parallel as long as each gate provides a distinct optical readout. A sufficiently high density of nanoparticles is maintained to ensure that a large number of logic-gated nanoparticle reactions can occur within a short period of time. More than approximately 4000 nanoparticles (>3700 receptors and 300 floaters) are tethered to an area of 180 × 180 μm2 for computing processes that typically last 15 to 30 min (fig. S5). We set the number of receptors to be higher than that of floaters to minimize trimer and tetramer formation. This condition allows the floaters to switch exclusively between the monomer and dimer states.
To rapidly and reliably process the dark-field imaging data, we introduce an image analysis pipeline that enables detection, tracking, and visualization of nanoparticle signals (Fig. 1D and fig. S6). The key to the analysis pipeline is our single-particle tracking algorithm that is capable of identifying and tracing signals from receptor particles in the high-density setting. Using the single-particle tracking algorithm, we profile the scattering signals of the receptors from a raw dark-field image sequence and generate a new movie whose each frame visualizes only the receptor signals in a dark background. This process allows the receptor signals to be clearly segmented from the background and distinguished from each other, providing a clear view of how nanoparticle circuits operate at the single-particle level in real time (movie S3). During experiments, we used this receptor-only visualization to qualitatively estimate the overall computing performance of a nanoparticle logic gate. We also apply the algorithm to profiling the signals of the red, green, and blue core nanoparticles. The signal profile, where single-particle signals of the three core particles are visualized in 3D space of a red-green-blue (RGB) scatterplot, shows three distinct red, green, and blue signal clusters (fig. S7). We use the 3D signal profile to filter out ambiguous signals that do not fall in the three signal clusters from subsequent analysis.
To quantitatively analyze the computing performance of nanoparticle logic gates, we perform kinetics analysis. By cumulatively counting the number of receptors that correctly exhibit monomer-to-dimer (for Assembly gates) or dimer-to-monomer (for Disassembly gates) transitions over time, we obtain the kinetics plots of assembly or disassembly outputs. The time-versus-output plots provide quantitative information on how accurately and fast logic gates respond to different combinations of molecular inputs. For example, the performance of an Assembly YES gate responding to two different input conditions is shown in Fig. 1E. As shown in the dark-field snapshots and the plot, the number of dimerization events between G-NRs and G-NF increases over time only in the presence of the input Xa (input = 1). This result indicates that the population of the nanoparticle logic gate switches into the ON state (output = 1) in response to Xa, performing a YES logic operation. To accurately perform computations, a population of nanoparticle logic gates should produce high output counts only when the molecular inputs meet TRUE conditions.
Design and construction of single-nanoparticle logic gates
We first demonstrated four types of two-input single-nanoparticle logic gates: Assembly AND, Assembly OR, Disassembly AND, and Disassembly OR gates. To construct these gates, we programmed receptor–floater interfaces in such a way that DNA bonds at the interfaces are formed (via assembly) or cleaved (via disassembly) only if two different DNA inputs satisfy AND or OR logic. We termed this approach “interface programming.” In the two-input Assembly AND gate, conformation-switchable DNA hairpins are used as surface ligands; a G-NR (R1) and a G-NF (F1) are each modified with a 5′-thiolated DNA hairpin that hides its binding domain in the stem (Fig. 2A and fig. S8A). Through hybridization with an input strand on its loop domain (t1 on F1; t2 on R1), the hairpin is opened and exposes the binding domain (b1*-a1* on F1; a1-b1 on R1). R1 and F1 assemble only if X1 and X2 are both present in the solution (movie S4). In the two-input Assembly OR gate, a G-NR (R2) and a B-NF (F2) are each modified with two types of DNA ligands; each particle provides two distinct binding domains (a2* and a3* on F2; b2* and b3* on R2) (Fig. 2B and fig. S8B). Either X3 or X4 can hybridize with the binding domains on F2 and R2, resulting in dimerization (movie S5). In the two-input Disassembly AND gate, a G-NF (F3) is initially bound to a B-NR (R3) by two different DNA bonds, each of which exposes a toehold domain at the R3–F3 interface (Fig. 2C and fig. S8C). The toehold domains t5 and t6 both act as recognition regions, recruiting input strands X5 and X6, respectively. Each input removes a DNA bond via strand displacement. When the two DNA bonds are all removed by X5 and X6, the disassembly reaction is initiated (movie S6). In the two-input Disassembly OR gate, a B-NF (F4) is bound to a B-NR (R4) via a DNA bond that exposes two toehold domains t7 and t8 at the interface, and either of the two domains can independently recruit an input strand X7 or X8 (Fig. 2D and fig. S8D). This design enables each input strand, whose sequence domains are complementary to half of the preformed DNA bond, to cleave the bond through strand displacement. Thus, either X7 or X8 induces the release of F4 (movie S7). The design principles for the interface programming are straightforward and generalizable (fig. S9).
Fig. 2. Nanoparticle logic gates.
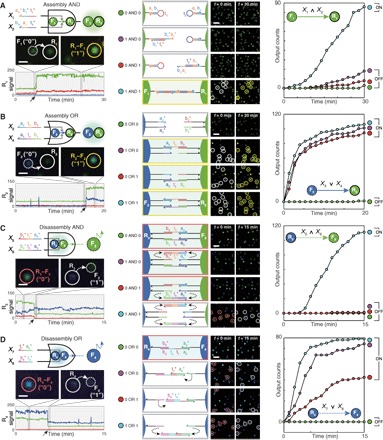
(A) Two-input Assembly AND. (B) Two-input Assembly OR. Assembly of G-NFs (F1) onto G-NRs (R1) and that of B-NFs (F2) onto G-NRs (R2) were traced as outputs for the AND and OR gates, respectively. (C) Two-input Disassembly AND. (D) Two-input Disassembly OR. Releases of G-NFs (F3) and B-NFs (F4) were traced as outputs for the Disassembly AND and OR gates, respectively. Each nanoparticle species is distinguished on the basis of its signal (color), mobility (R or F), and function (subscripted number). Domains a1t* and b1t [3 nucleotides (nt)] are truncated versions of a1* and b1 (6 nt), respectively. First column: Circuit diagram (top), actions of a single-nanoparticle logic gate in a logical TRUE condition (middle), and the time trace of a receptor signal (bottom). Solid and dashed arrows in the x axis indicate monomer-to-dimer and dimer-to-monomer transitions of receptors, respectively. Scale bars, 1 μm. Second column: Domain-level illustrations (left) and population-level responses (right) of the logic gates. Scale bars, 4 μm. Receptors are marked with white circles before assembly (red circles before disassembly) and yellow circles after assembly (gray circles after disassembly). Third column: Kinetics analysis. Each plot contains a reaction graph that corresponds to a logic gate. ∧ and ∨ denote the logic symbols for AND and OR, respectively. Legends are represented as truth tables. DNA sequences and experimental conditions are listed in tables S2 and S3. Experiments were carried out at 25°C in 1× phosphate-buffered saline (PBS) buffer.
We monitored the operations of the four elementary logic gates in real time using DFM. Single-particle snapshots of each logic gate responding to a logical TRUE condition (1 AND 1 for AND gates and 1 OR 1 for OR gates) are provided in the first column of Fig. 2. The representative time traces of the receptor-scattering signals show that each two-input gate exhibits a distinct optical readout. The change in a receptor signal is largely determined by a floater signal: Reactions with G-NFs result in an increase or a decrease in green scattering intensity of receptors (shown in Fig. 2, A and C), and reactions with B-NFs lead to changes in both green and blue intensities (shown in Fig. 2, B and D). As the four logic gates all produce spectrally distinct signals, we can, in principle, operate the four gates simultaneously. Furthermore, operations of the logic gates captured in a large field of view are shown in the third column of Fig. 2. The images were processed to visualize only receptor signals. For each logic gate, we can readily determine whether the logic gate responds to molecular inputs by simply comparing the first and last frames of a processed movie.
We quantified the kinetics of the two-input logic gates by counting their particle-by-particle responses over time. All four logic gates—Assembly AND, Assembly OR, Disassembly AND, and Disassembly OR—generated low output counts under the logical FALSE conditions and high output counts under the TRUE conditions, providing ON/OFF levels over 5 folds, 88 folds, 93 folds, and 42 folds with fast response kinetics (t1/2 < 19 min, t1/2 < 5 min, t1/2 < 9 min, and t1/2 < 5 min), respectively. We calculated ON/OFF levels by dividing the lowest output count obtained in TRUE conditions by the highest output count obtained in FALSE conditions. The t1/2 of a nanoparticle logic gate was obtained by measuring the amount of time needed for half of its floaters to correctly respond to molecular inputs in TRUE conditions. The response rate (%), defined as the number of floaters that react to inputs divided by the total number of floaters, was ~80% in TRUE conditions (table S1). Given that ~10% of floaters inevitably participate in multimer formations (fig. S5D), the measured response rates suggest that more than 90% of floaters eventually exhibited state-switching behaviors. The Assembly AND gate generated a small number of incorrect output signals at 1 AND 0 and 0 AND 1 conditions presumably because the surface hairpins are in dynamic equilibrium between closed and opened states. Despite the minor leakage, the hairpin-based particle assembly is exclusively driven by input-induced hairpin opening (fig. S10A) and occurs without interfering with other hybridization events (fig. S10B). The two-input Disassembly OR gate exhibited uneven responses, where the 0 OR 1 condition results in a response rate of 42% that is notably lower than those of other two TRUE conditions (74% in the 1 OR 0 condition and 79% in the 1 OR 1 condition). We attribute this result to the difference in the density of surface ligands between receptors and floaters (fig. S11). In addition, we showed that the computing architecture of LNTs is compatible with a “dual-rail” convention, where the Boolean values of a logic gate are represented by the presence of either one signal (“0”) or another (“1”). This formalism is frequently used for systems where it is difficult to directly introduce NOT function (8, 10, 12). With this representation, AND and OR gates are sufficient to compute any Boolean functions. We demonstrated a two-input dual-rail NAND gate as a proof of concept (fig. S12).
Next, the interface programming approach is expanded to enable nanoparticle logic gates to process INHIBIT logic [X1 AND (NOT X2)], take multiple inputs (fan-in), and generate multiple outputs (fan-out). First, we implemented a two-input Disassembly INHIBIT gate (Fig. 3A). To realize the NOT logic required for the INHIBIT gate, we exploit competition between DNA bond elimination (triggered by X1) and formation (triggered by X2). The INHIBIT gate releases a G-NF (F1) as an output, only when the disassembly input X1 is present and the assembly input X2 is absent (fig. S13A and movie S8). The INHIBIT gate generated outputs only in the TRUE states with an ON/OFF level over 129 folds (t1/2 < 10 min). No output leakage was observed when both inputs were present, indicating that the bond formation by X2 is faster than the bond removal by X1. In addition, the two competing reactions proceed without interfering with each other (fig. S13B). Demonstration of the INHIBIT gate is important because two-input AND, OR, and INHIBIT operations constitute a functionally complete set of Boolean functions. Second, we constructed a multi-input Disassembly gate by increasing the number of distinct DNA bonds in a receptor–floater dimer. When the release of a G-NF (F2) from a G-NR (R2) requires the disconnection of three different DNA bonds and each disconnection is controlled by two-input OR logic, the disassembly is regulated by a six-input expression (X3 OR X4) AND (X5 OR X6) AND (X7 OR X8) (Fig. 3B). Representative domain-level illustration and dark-field snapshots of the six-input logic gate responding to a FALSE condition [(0 OR 0) AND (0 OR 0) AND (0 OR 0)] and a TRUE condition [(1 OR 0) AND (1 OR 0) AND (1 OR 0)] are provided. Dark-field time-lapse imaging confirmed that the six-input logic gate produced outputs only in the TRUE states with an ON/OFF level over 88 folds (t1/2 < 9 min) (fig. S13C and movie S9). The two strategies based on competing reactions and increased “bond orders” can be combined to yield Disassembly gates with complex multi-input Boolean logic, such as (X1 OR X2) AND [NOT (X3 OR X4)] and (X1 AND X2) AND (NOT X3) (fig. S14). Third, we demonstrated the fan-out of a logic gate by implementing identical two-input Disassembly AND logic in three different receptor–floater pairs, each of which has a spectrally distinct floater signal (Fig. 3C). The disassembly reactions of the three floaters were readily analyzed, owing to plasmonic coupling between the receptors and the floaters; the dissociations of R-NF (F3), G-NF (F4), and B-NF (F5) from the receptors resulted in decreases in red, green, and blue intensities of the receptor signals, respectively (movie S10). The Disassembly gate with fan-out released all three outputs according to the AND logic, with ON/OFF levels over 20 folds (t1/2 < 6 min).
Fig. 3. Circuits with an INHIBIT logic gate, multiple inputs (fan-in), or multiple outputs (fan-out).
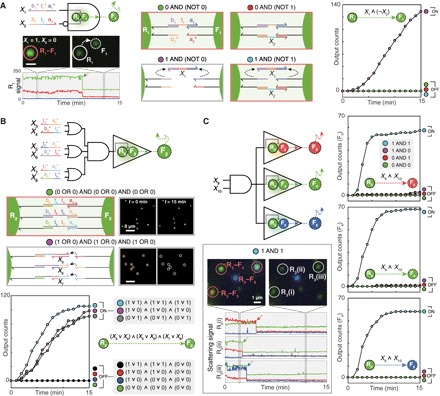
(A) Two-input Disassembly INHIBIT gate implemented in a pair of G-NR (R1) and G-NF (F1). X1 removes a preformed DNA bond via strand displacement, whereas X2 forms a different DNA bond. Disassembly of the R1–F1 dimer is controlled by INHIBIT logic (X1 AND NOT X2) based on the two competing reactions. Actions of single-particle INHIBIT gate are shown. Scale bar, 1 μm. (B) Six-input Disassembly gate implemented in a pair of G-NF (R2) and G-NF (F2). The receptor (R2) and the floater (F2) are connected by three distinct DNA bonds, and the cleavage of each bond is controlled by two-input OR logic. (C) Two-input Disassembly AND gate with three outputs. This circuit evaluates two-input AND logic and generates three different mobile floaters—R-NF (F3), G-NF (F4), and B-NF (F5)—as outputs. Three representative receptors that release F3, F4, and F5 are labeled as R3(i), R3(ii), and R3(iii), respectively. Dashed arrows in time traces indicate disassembly reactions. The range of y-axis values in the time traces is 0 to 250. ∧, ∨, and ¬ denote the logic symbols for AND, OR, and NOT, respectively. Legends are represented as truth tables. DNA sequences and experimental conditions are listed in tables S2 and S3. Experiments were carried out at 25°C in 1× PBS buffer.
Modular wiring of nanoparticle logic gates into a circuit
As the complexity of reactions at the receptor–floater interface increases, incomplete reactions or spurious interactions arise as well. Thus, relying on the programming of particle interfaces is not a scalable strategy for constructing complex circuits. Instead, we introduce a conceptually distinct approach—the nanoparticle “network programming”—that enables any two single-particle logic gates to be combined with AND or OR logic. According to nanoparticle reaction graph abstraction, a nanoparticle logic gate is represented by a graph composed of two nodes and one edge. Through network programming, two edges, each of which represents an assembly or disassembly reaction, are connected by a floater node. This process is equivalent to the coupling of two nanoparticle reactions and thus enables more complex logic operations to be implemented at the level of the nanoparticle network. Joining a Disassembly edge (Gate 1) with an Assembly edge (Gate 2) represents the wiring of the two gates with AND logic, and linking two Disassembly edges (Gates 3 and 4) symbolizes the wiring of the two gates with OR logic.
First, we demonstrated the wiring with AND logic by allowing a floater to be used in a Disassembly gate and an Assembly gate in series. We designed a two-input Disassembly AND gate composed of a G-NR (R1) and a G-NF (F1) in such a way that the F1 acts as an Assembly YES gate with another receptor B-NR (R2) after dissociation from the R1 (Fig. 4A). In this network-level wiring scheme, the formation of an R2–F1 dimer then becomes an output of the AND-AND cascade circuit (X1 AND X2) AND X3 (fig. S15A and movie S11). The circuit is described by a reaction graph, where two receptors (R1 and R2) are serially connected to a floater (F1). Dark-field imaging revealed that the circuit provided an ON/OFF level of 36 folds. The intermediate disassembly reactions could also be analyzed owing to their distinct optical signals. The upstream Disassembly AND gate resulted in an ON/OFF level over 89 folds. Furthermore, we quantified how F1 population changes over time by subtracting the number of receptors from the total number of detected nanoparticles in each frame. The population dynamics of F1 shows that the released F1 subsequently binds to R2 only if the input X3 is present. For (1 AND 1) AND 1 condition, more than 92% of released F1 responded to the assembly input X3. This result indicates that the sequential disassembly-assembly cascade is highly efficient. In addition, the accumulation of F1 population in (1 AND 1) AND 1 indicates that the assembly reaction is relatively slower than the disassembly reaction in this case. The reversal of the reaction kinetics observed in this condition arises presumably because the access of input X3 is temporarily hindered by DNA strands hybridized on the floater. Other two-input Disassembly gates, such as two-input OR and INHIBIT gates, could be modularly rewired, resulting in an OR-AND cascade circuit (fig. S16A) and an INHIBIT-AND cascade circuit (fig. S16B). Second, we implemented OR wiring by designing two Disassembly gates to produce floaters with identical optical signals (Fig. 4B). We installed a two-input Disassembly AND gate [composed of a B-NR (R3) and a G-NF (F2)] and a Disassembly YES gate [composed of a G-NR (R4) and another G-NF (F3)] in parallel. In the circuit (X4 AND X5) OR X6, G-NFs can be produced from either the AND gate or the YES gate; the two gates are thus wired with OR logic (fig. S15B and movie S12). The AND-OR cascade circuit resulted in an ON/OFF output level of 55 folds. We also evaluated the operations of the two upstream Disassembly gates separately, showing that the two gates carried out computation with high ON/OFF levels without interfering with each other. A two-input Disassembly OR gate can be also wired with a Disassembly YES gate to yield an OR-OR cascade circuit (fig. S17). Together, our results indicate that the network programming strategy can be readily applied to implementing complex multilayer cascades without extensive optimization.
Fig. 4. Wiring of nanoparticle logic gates via network programming.
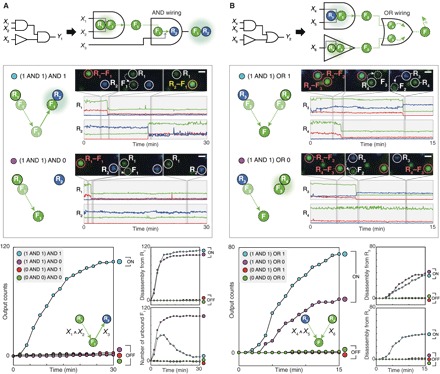
(A) Wiring with AND logic. Two logic gates (Disassembly AND gate and Assembly YES gate) are designed to operate in series for AND wiring. The floater F1, which is bound to the first receptor R1 in its initial state, acts as a Disassembly AND logic gate and subsequently as an Assembly YES gate with the second receptor R2. The generation of R2–F1 dimers is an output of the (X1 AND X2) AND X3 circuit. (B) Wiring with OR logic. Two logic gates (Disassembly AND gate and Disassembly YES gate) are designed to operate in parallel for OR wiring. The two gates both release G-NFs as outputs. The generation of the G-NFs is an output of the (X3 AND X4) OR X5 circuit. Circuit diagrams (top), single-particle dark-field analysis (middle), and kinetics analysis of circuits (lower left) and intermediate reactions (lower right). Scale bars, 1 μm. Legends are used as truth tables. DNA sequences and experimental conditions are listed in tables S2 and S3. Experiments were carried out at 25°C in 1× PBS buffer.
A nanoparticle multiplexer circuit on a lipid bilayer
To demonstrate the modularity of nanoparticle computing on the LNT platform, we implemented a multiplexer circuit (MUX2to1) by rewiring previously introduced logic gates via network programming. The multiplexer takes a selector input (Sel) to select one of two inputs X1 and X2 and translates the selected input into a single output (Fig. 5A). The multiplexer circuit is designed by connecting a Disassembly INHIBIT gate (X1 AND NOT Sel) and a two-input Disassembly AND gate (Sel AND X2) with OR logic. This circuit diagram is translated into a reaction graph diagram that subsequently guides the design of surface DNA ligands for each nanoparticle. A multiplexer circuit is composed of two receptor–floater pairs, R1–F1 and R2–F2, whose interfaces can evaluate (X1 AND NOT Sel) and (Sel AND X2) operations, respectively. Both producing G-NFs as outputs, the two Disassembly gates are wired with OR logic. The multiplexer circuit requires identical selector inputs to be simultaneously processed by two different logic operations: INHIBIT logic (by R1–F1 pair) and AND logic (by R2–F2 pair) (fig. S18). Under this design constraint, F1 and R1 should expose sequence domains a2 and b2 that are fully complementary to surface ligands on F2 and R2, respectively (Fig. 5B). In solution phase, these nanoparticles would spontaneously form aggregates; F1 can bind to F2 through complementary binding domains a2 and a2*. The multiplexer circuit, however, could be constructed on a lipid bilayer without such issues because the undesirable, spontaneous interactions between the nanoparticles are compartmentalized and controlled during circuit preparation steps: F1–F2 interaction is inhibited by temporary protection of the a2* domain (introduced before the tethering of F1), and R1 and R2 do not collide with one another because of their immobility on SLBs. After the four circuit components are all loaded on the lipid bilayer, two Disassembly logic gates are prepared by forming the R1–F1 and R2–F2 dimers. As shown in Fig. 5C and movie S13, the nanoparticle multiplexer yielded the expected responses to the eight different input combinations with an ON/OFF level over 35 folds. The output counts of the multiplexer come from either a Disassembly INHIBIT gate (R1–F1) or a Disassembly AND gate (R2–F2). In logical TRUE conditions, the circuit exhibited uneven responses because the INHIBIT gate, which requires the removal of only one DNA bond (X1*), is kinetically more favorable to disassemble than the Disassembly AND gate that requires the simultaneous removal of two different DNA bonds. The successful implementation of the nanoparticle multiplexer shows that one can design and operate nanoparticle circuits on LNTs in a highly modular and controllable manner.
Fig. 5. A nanoparticle multiplexer circuit.
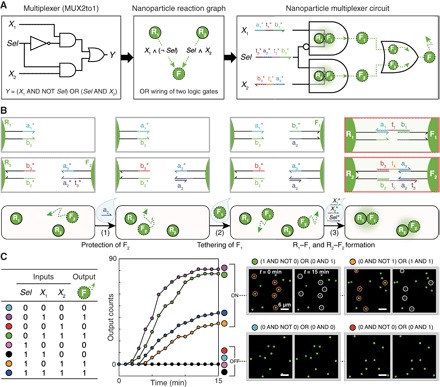
(A) Multiplexer circuit (MUX2to1) implemented by a network of nanoparticles. The multiplexer is constructed by wiring a Disassembly INHIBIT gate (X1 AND NOT Sel) and a Disassembly AND gate (Sel AND X2) with OR logic. Four green nanoparticles (R1, R2, F1, and F2) constitute the multiplexer circuit. (B) Modular construction of the nanoparticle multiplexer on a lipid bilayer. The circuit components could be loaded in a controlled and modular manner. To prevent unwanted spontaneous interaction between F1 and F2, F2 was hybridized with the protector strand a2 before loading F1. (C) Measured performance of the multiplexer circuit. Truth table (left), kinetics analysis (middle), and dark-field snapshots (right). Domain-level illustration of the circuit operation is illustrated in fig. S18. Scale bars, 4 μm. DNA sequences and experimental conditions are listed in tables S2 and S3. Experiments were carried out at 25°C in 1× PBS buffer.
DISCUSSION
The uniqueness of LNTs lies in the following three merits. First, the computation is solely driven by SLB-tethered nanoparticles whose particle-by-particle interactions are programmable and readable in situ. A dynamic network of individual nanoparticles thus acts as a logic circuit. Second, information relay in the dynamic nanoparticle circuits does not require signal restoration or amplification because the cascading is exclusively driven by mobile floaters. In this context, floaters are “wires” carrying the information of upstream gates into downstream gates via their robust lateral diffusion (fig. S19). Third, spatial constraints are exploited to direct the information flow in dynamic-yet-confined nanoparticle networks. Complex digital logic operations can be implemented using a relatively small number of particle and ligand types with few design constraints, as reaction centers can be separated by immobile receptors. The demonstration of the multiplexer circuit exemplifies the beneficial role of spatial constraints in the LNT platform. Through the integration with a lipid bilayer, nanoparticles exhibit new collective properties—digital principles—that are absent and cannot be realized in their static, solution-phase assemblies.
The scope of lipid bilayer–based nanoparticle computation can be expanded to advance current molecular computing technologies. First, solution-phase molecular circuits that generate single-stranded DNA as outputs (7, 8, 13, 14) can be synergistically interfaced with LNTs because the released DNA molecules can be used as new inputs to operate the nanoparticle circuits. Through the additional layer of molecular circuitry in solution, different nanoparticle circuit modules on a lipid bilayer can communicate with each other. Second, particle modifications based on chemical ligands other than DNA can be introduced to implement nanoparticle circuits that process diverse chemical information (37). When nanoparticles are functionalized with new chemical ligands, the design constraints (that may arise from the crosstalk between different surface ligands) can be reduced because particle-by-particle interactions can be spatiotemporally controlled on a lipid bilayer. Third, integrations of lipid bilayers with DNA nanostructures (38) may provide a path toward the development of new molecular circuits. If DNA origami scaffolds that contain spatially localized DNA circuits are tethered to SLBs, one may be able to exploit a dynamic network of inter-origami interactions to implement more complex and practical molecular computation.
Despite such potentials, further scaling up the complexity of nanoparticle circuits on LNTs will be challenging because the input (molecules in solution) and output (state-switching floaters) are of different forms. Currently, this intrinsic difference limits the construction of arbitrarily large circuits. We predict that this challenge can be potentially addressed in two ways. First, harnessing new modes of nanoparticle reaction and ligand activation—such as communication (3, 4), dynamic reconfiguration (39), and DNA walker (40)—may provide a much broader design space for circuit design. Second, increasing the number of different nanoparticle computing units per lipid bilayer “computing chip” will enhance the processing power because these units can operate as a complex circuit through network programming. This approach is similar to the way silicon-based computers have improved over the years: An increase in component density enhances their computing capacity. Exploiting the potential of parallelism, we will ultimately have each nanoparticle independently perform a computation on its own.
As spatial constraints such as localization and encapsulation enable the modular execution of molecular and synthetic biological circuits (10, 41, 42), tethering of nanoparticles to a lipid bilayer provides a systematic method to build complex nanoparticle circuits. The LNT platform will play a pivotal role in constructing dynamic, “autonomous” nanosystems and devices, which will have broad impacts in the following areas. First, the LNT platform can be applied to molecular diagnostics and smart sensors; the systems of individual nano-objects in the devices should be able to exploit internal computational algorithms to sense multiple stimuli and trigger the most appropriate responses. Second, if the nanoparticle circuits are introduced to living cell membranes (43), it might be possible to create new types of nano-bio interfaces that are useful for cell-surface engineering and biological-inorganic hybrid systems. Lastly, information-processing nanoparticles on SLBs can be applied to reconstituting artificial cell-cell junctions and used as tools for studying membrane-associated phenomena in living cells. Unlike existing methods that rely on immutable materials such as patterned membranes (34), LNTs will allow networks of SLB-tethered nanostructures to algorithmically form clusters in response to signaling molecules released from the cells. Facilitating the nanosystem and cellular system to communicate with one another, such “active” SLB-cell junctions can also be used to test how individual theranostic nanorobots navigate complex and dynamic environments.
MATERIALS AND METHODS
To construct LNTs, three key components—small unilamellar vesicles (SUVs), glass flow chambers, and DNA-functionalized plasmonic nanoparticles—were first prepared. Afterward, the solution of SUVs was introduced into a flow chamber to form an SLB on its bottom glass substrate. The DNA-modified nanoparticles were then tethered to the SLB and used as logic gates and circuits for molecular information processing. Depending on their mobility on an LNT, the functionalized nanoparticles were classified into immobile receptors (reporters of computation) or mobile floaters (information carriers of computation). Receptors and floaters for Disassembly logic gates were predimerized before each logic operation. DFM imaging was carried out to measure the performances of the nanoparticle logic gates responding to molecular inputs in solution. Dark-field image sequences obtained from the logic operations were processed and quantified by a custom-built image analysis pipeline.
Preparation of SUVs
The lipid solution in chloroform was mixed in a round-bottom flask to have 97.2 mole percent (mol %) dioleoylphosphatidylcholine, 0.3 mol % biotinylated dioleoylphosphatidylethanolamine (DOPE), and 2.5 mol % poly(ethylene glycol) 1000 (PEG 1000)–DOPE. Chloroform was removed using a rotary evaporator, and the lipid film formed inside the flask was thoroughly dried for 15 min under a stream of N2. The dried mixture was resuspended in deionized (DI) water to have a total concentration of 2 mg/ml. The lipid solution went through three freeze-thaw cycles between −78° and 40°C. To generate uniform SUVs, the solution was extruded more than 11 times through a polycarbonate membrane with a pore diameter of 100 nm at 30°C. The resulting SUV solution was kept at 4°C until use.
Preparation of SLBs
SLBs were prepared on glass substrates within flow chambers via the vesicle fusion method. A flow chamber was made from a top and bottom glass and a Parafilm spacer (4 mm × 50 mm × 200 μm). The working volume of the glass chamber is ~40 μl. The top slide glass (Paul Marienfeld GmbH & Co. KG) with inlet and outlet holes was cleaned by 5-min bath sonication in DI water and 2-min piranha etching in H2SO4/H2O2 (3:1). After each cleaning procedure, glass substrates were rinsed with sufficient amounts of DI water. The cleaned top glass slide was then passivated with bovine serum albumin (BSA) (10 mg/ml) in 150 mM NaCl phosphate-buffered saline (1× PBS) to prevent SLB formation on the upper side of the chamber. The bottom cover glass (Paul Marienfeld GmbH & Co. KG, Germany) was cleaned by 5-min sonication in DI water followed by 2-min piranha etching. A two-ply Parafilm spacer was then placed between the two glass slides and heat-sealed at 100°C. The freshly extruded SUV solution was diluted to 1 mg/ml in 1× PBS solution and sonicated for 15 min. The SLBs were formed by introducing the sonicated vesicle solution into the flow chamber at 30°C. After 60 min, the flow chamber was gently washed with DI water and 1× PBS. Defects in SLBs were then passivated with BSA (100 μg/ml) in 1× PBS for 45 min. Streptavidin (17 nM) in 1× PBS was then injected into the flow chamber to modify the biotinylated DOPE molecules. After 40 min, the flow chamber was washed with 1× PBS twice. The flow chamber with the streptavidin-modified SLB can be stored up to 3 days in a humidified refrigerator at 4°C. Formation of air bubbles inside the chamber should be avoided in all procedures.
Synthesis and functionalization of plasmonic nanoparticles
Gold nanorods with silver shells, gold nanospheres, and silver nanospheres on gold seeds that exhibit red, green, and blue scattering signals were synthesized and referred to as red, green, and blue nanoparticles, respectively. For the preparation of red nanoparticles, gold nanorods with an aspect ratio of 4 were first synthesized according to previous methods based on a seed-mediated growth mechanism (36). Silver shells (5 nm thick) were formed around the gold nanorods by incubating the gold nanorod solution (1 ml, 100 nM) with cetyltrimethylammonium chloride solution (1 ml, 10 mM), AgNO3 (1 ml, 0.2 mM), and l-ascorbic acid (1 ml, 50 mM) for 4 hours. The resulting red nanoparticles (diameter, ~22 nm; length, ~56 nm) were washed three times by centrifugation, supernatant removal, and redispersion in DI water. Spherical gold nanoparticles (diameter, ~50 nm) were purchased from BBI Solutions (Cardiff, UK) and used as green nanoparticles. Blue nanoparticles (diameter, ~54 nm) were prepared by growing 17-nm-thick silver shells on 20-nm spherical gold nanoparticle seeds. To form silver shells on the seeds, sodium ascorbate solution (100 μl, 50 mM) was rapidly injected into the mixture containing 150 pM of 20-nm gold seeds, 0.2% polyvinylpyrrolidone (PVP), and 0.24 mM AgNO3. The nanoparticles were characterized by transmission electron microscopy (TEM; JEM-2100, JEOL Ltd., Japan), UV-Vis spectrophotometry (Agilent 8453, Agilent Technologies, USA), DFM (Axiovert 200 M, Carl Zeiss, Göttingen, Germany), and field-emission scanning electron microscopy (FE-SEM; JSM-7600F, JEOL Ltd., Japan). Correlative DFM-SEM imaging was performed to analyze single-particle scattering signals from the three nanoparticles: Nanoparticles loaded on a Cr-patterned glass substrate were first imaged by DFM, treated with Pt coating (Cressington 108auto, Cressington Scientific Instruments Ltd., UK), and imaged in the same position by FE-SEM. The characterization data are summarized in fig. S2. TEM and FE-SEM imaging were carried out at the National Center for Inter-University Research Facilities and at the Research Institute of Advanced Materials (both at Seoul National University, Seoul, South Korea), respectively.
Thiolated DNA oligonucleotides (Bioneer, Daejeon, Korea) were treated with 100 mM dithiothreitol in 100 mM phosphate buffer (PB) (pH 8.0) for 1 hour. Afterward, the oligonucleotides were purified via size-exclusion chromatography with a NAP-5 column (GE Healthcare, Buckinghamshire, UK). Nanoparticles (final concentration of 15 pM) were incubated with 216 nM (for blue nanoparticles) or 288 nM (for red and green nanoparticles) thiolated oligonucleotides for 1 hour at 25°C. The ratios of biotinylated DNA linkers to total surface DNA ligands were 0.5, 0.5, 0.5, 35, 35, and 50% for R-NF, G-NF, B-NF, R-NR, G-NR, and B-NR, respectively. The solution was then adjusted to 0.1% (w/v) PVP in 10 mM PB for red nanoparticles, to 0.1% (w/v) SDS in 10 mM PB for green nanoparticles, and to 10 mM PB for blue nanoparticles. Three aliquots of 1 M NaCl, 0.1% SDS, and 10 mM PB salt solution were added with a 1-hour interval to achieve a final concentration of 0.3 M NaCl. After each salt addition, the mixture was heated at 50°C for 10 min and incubated at 25°C for 50 min. Two hours after reaching the final concentration, the red nanoparticle solution was centrifuge-washed and redispersed in 1× PBS. Other nanoparticle solutions were incubated for 12 hours, centrifuge-washed, and redispersed in DI water (green nanoparticles) or in 1× PBS (blue nanoparticles).
Oligonucleotide design
Nanoparticles were functionalized with single-stranded DNA strands that contain thiol modifications at their 3′ or 5′ ends. DomainDesign (44, 45) was used to generate a set of orthogonal 10-nucleotide (nt) toeholds, 10-nt supporting domains, 14-nt binding domains, and 34-nt linker domains. These sequences were then verified using NUPACK (46) and further optimized if any undesirable interaction was detected during experiments. The thiolated DNA strands include (i) biotinylated linker DNA strands used for the tethering of nanoparticles to streptavidin-modified SLB surfaces and (ii) ligand DNA strands directly involved in the nanoparticle computing process. A linker strand with 5′-thiol modification contains (i) an A15 domain after a 5′-thiol group, (ii) six ethylene glycol (EG) units (a PEG moiety), and (iii) a linker domain followed by biotin modification. A linker strand with 3′-thiol modification (with biotin modification at 5′ end) contains (i) a linker domain, (ii) a PEG moiety, and (iii) an A15 domain followed by 3′-thiol modification. Ligand DNA strands were categorized into two types: “normal” single-stranded DNA strands that do not form hairpin loops and hairpin-type DNA ligands used in Assembly AND gate. The normal ligand type was further classified into two groups: one with 3′-thiol modification and another with 5′-thiol modification. A ligand with 5′-thiol modification contains (i) an A15 domain after 5′-thiol, (ii) a PEG moiety, (iii) a supporting domain, and (iv) a binding domain. A ligand with 3′-thiol modification contains (i) a binding domain, (ii) a supporting domain, (iii) a PEG moiety, and (iv) an A15 domain followed by 3′-thiol modification. Unless otherwise noted, receptor and floaters were functionalized with 3′ thiol ligands and 5′ thiol ligands. Hairpin-type DNA ligands were thiolated at 5′ end. Supporting domains were introduced to better expose binding domains to solution, thereby promoting the hybridization of the binding domains with input strands. The A15 domain and PEG moiety in each strand are essential parts of the design, as these two components enhance DNA hybridization on nanoparticle surfaces and decouple functional units (binding domains) from a core structure (nanoparticle) by providing optimal DNA density, reducing nonspecific interactions, and stretching out the binding domain. Sequences of thiolated strands are listed in table S2.
Operation and characterization of nanoparticle circuits on LNTs
A solution containing ~3 pM of DNA-modified nanoparticles with biotinylated linkers was introduced to a flow chamber, whose bottom glass substrate was coated with a streptavidin-modified lipid bilayer. The solution was incubated from 1 to 5 min to result in a desired particle density. The particle density was linearly proportional to incubation time (fig. S5). After the particle loading, the LNT was washed with 1× PBS twice. The tethered nanoparticles then functioned as logic gates, taking single-stranded DNA inputs in 1× PBS buffer. The performance of each nanoparticle circuit was tested by injecting 500 μl of an input solution into the flow chamber during dark-field imaging. The input solution was injected slowly while a user was monitoring the field of view such that a flow introduced from the injection did not alter the initial position of receptors. Dark-field imaging was carried out at room temperature by a commercial dark-field microscope with a 40× objective lens (numerical aperture, 0.6) and AxioCam HRc color camera on an optical table (Daeil Systems, South Korea). The field of view was 180 × 180 μm2. Before input injection, 31 images were acquired with a time step of 200 ms to identify the positions and signals of receptor nanoparticles. Circuit performance was recorded during and after input injection with an imaging interval of 2.5 s. The recording continued until the responses of the circuits reached a plateau. Both image sequences were acquired at a fixed position. Sequences of input strands and experimental conditions for logic gate operation are listed in table S3.
Analysis of dark-field time-lapse images
Images obtained from time-lapse dark-field imaging were analyzed using custom MATLAB code to quantify how nanoparticle circuits respond to molecular inputs. Images were first registered using StackReg plugin in ImageJ. To avoid sampling bias resulting from uneven focus and illumination across the field of view (that were typically observed along the image boundaries), an area of interest (128 × 128 μm2) was then chosen for the analysis. Afterward, the drift-corrected image sequences were processed by an image analysis algorithm that enables single-particle tracking and signal classification. The single-particle tracking algorithm consists of the following three steps: (i) In the signal detection step, spots (pixels) with signals considerably higher than threshold intensity were identified using a pixel-based intensity threshold. The contours obtained from the detected pixels enabled the segmentation of each nanoparticle signal. (ii) In the particle localization step, the representative position of each segmented signal was determined. Each localized nanoparticle was assigned with an intensity value obtained from averaging pixel intensities from 3 pixels × 3 pixels around the localized position. (iii) In the particle tracking step, nanoparticle signals, whose positions remained unchanged for the entire imaging duration, were identified as receptor signals. Time traces of all receptor signals identified in the chosen field of view were obtained and used as a signal profile of each image sequence. The signal profile was then used to generate a receptor-only image sequence. The algorithm is described in detail in fig. S6. In classification step, only the receptors that exhibit step-function–like signal traces were identified as output-generating particles. Transient interactions (short, sharp increase or decrease of a receptor signal) that resulted from temporary colocalizations of two nanoparticles in diffraction-limited spots were filtered out. Any intensity changes that do not last until the end of imaging were excluded. To reliably categorize the nanoparticle reaction types, a signal profile of red, green, and blue nanoparticles shown in fig. S7 was used as a standard. Ambiguous signals that did not fall in the red, green, and blue signal clusters in the standard signal profile were excluded. Afterward, RGB scattering signals of the receptor were further analyzed to identify the floater with which the identified receptor reacted. The entire analysis process is operated in a MATLAB-based graphical user interface program (fig. S6D).
For each logic gate, a time-versus-output relationship was plotted to reveal accuracy and kinetics of the logic gate computation. Output counts of the plot were obtained by cumulatively counting the number of receptors that generated correct outputs over time. To minimize the effect by the variability in particle populations, the final output counts were normalized for each type of logic gate. For example, the output counts measured for the analysis of a two-input AND gate were normalized across the four input conditions (0 AND 0, 0 AND 1, 1 AND 0, and 1 AND 1). For Assembly gates, output counts were normalized by the number of floaters detected in the initial 31 images. For Disassembly gates, output counts were normalized by the number of dimers formed in the predimerization step. ON/OFF levels were calculated by dividing the lowest output count obtained in TRUE conditions by the highest output count obtained in FALSE conditions. If the output count in FALSE conditions was 0, then the value was set to be 1.
Diffusion behaviors of nanoparticles were analyzed as follows: (i) Particles were tethered to an SLB with a sufficiently low particle density (~200 particles per 180 × 180 μm2) that allowed long-term tracking (~10 min) without trajectory overlap. (ii) Nanoparticle signals from each frame were detected and localized by the image analysis algorithm. (iii) Determined positions were used to generate a trajectory of each particle, which was then used to calculate its diffusion coefficient. Mean square displacement (MSD) values as a function of time interval were obtained for each particle. The MSD plots of these trajectories were fitted to the equation, <r2> = 4Dt, where <r2> is the MSD, D is the diffusion coefficient, and t is the time interval.
Supplementary Material
Acknowledgments
We thank J.-E. Park and members of the Nam laboratory for discussions and comments on the manuscript. We also thank D. Lee and Z. H. Kim for discussions. Funding: This work was supported by Samsung Research Funding and Incubation Center of Samsung Electronics (SRFC-MA1502-02). J.S. acknowledges the Korea Foundation for Advanced Studies for undergraduate fellowship. Author contributions: J.S. and J.-M.N. conceived the original idea for the LNT platform. J.S., S.K., and H.H.P. designed and carried out the experiments under the guidance of J.-M.N. H.H.P. developed a software for analyzing dark-field image data under the guidance of J.-M.N. J.S. and H.H.P. analyzed the data with the guidance of J.-M.N. S.K. and D.Y.C. obtained TEM images. All authors discussed the results and commented on the manuscript. J.-M.N. supervised the project. J.S. and J.-M.N. wrote the manuscript with inputs from all authors. Competing interests: J.S., S.K., H.H.P., and J.-M.N. are inventors on a patent application related to this work filed by Seoul National University R&DB Foundation (patent application no. 10-2018-0061345, date: 29 May 2018). The authors declare no other competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/2/eaau2124/DC1
Fig. S1. Conceptual illustrations of the LNT platform.
Fig. S2. Characterization of nanoparticles.
Fig. S3. Diffusion of nanoparticles tethered to an SLB.
Fig. S4. Design and implementation of a Disassembly YES gate.
Fig. S5. Tethering of nanoparticles to an SLB.
Fig. S6. Image analysis pipeline.
Fig. S7. Scattering signal profiles of red, green, and blue nanoparticles.
Fig. S8. Nucleotide-level schematics of two-input logic gates.
Fig. S9. Design principles for nanoparticle logic gates.
Fig. S10. Modularity of a hairpin-based two-input Assembly AND gate.
Fig. S11. Uneven responses of a two-input Disassembly OR gate.
Fig. S12. A dual-rail NAND gate.
Fig. S13. Disassembly INHIBIT and six-input Disassembly gates.
Fig. S14. Complex multi-input Disassembly gates.
Fig. S15. Dark-field snapshots of two-layer AND-AND and AND-OR cascade circuits.
Fig. S16. Two-layer OR-AND and INHIBIT-AND cascade circuits.
Fig. S17. A two-layer OR-OR cascade circuit.
Fig. S18. A nanoparticle multiplexer circuit.
Fig. S19. Effect of DNA concentration on floater diffusion.
Table S1. Response rates of two-input logic gates.
Table S2. DNA sequences of thiolated strands used for functionalizing nanoparticles.
Table S3. DNA sequences and experimental conditions used in circuit operations.
Movie S1. Time-lapse dark-field imaging of a nanoparticle Assembly YES gate.
Movie S2. Time-lapse dark-field imaging of a nanoparticle Disassembly YES gate.
Movie S3. Receptor-only visualization of a dark-field movie.
Movie S4. Time-lapse dark-field imaging of a two-input Assembly AND gate.
Movie S5. Time-lapse dark-field imaging of a two-input Assembly OR gate.
Movie S6. Time-lapse dark-field imaging of a two-input Disassembly AND gate.
Movie S7. Time-lapse dark-field imaging of a two-input Disassembly OR gate.
Movie S8. Time-lapse dark-field imaging of a two-input Disassembly INHIBIT gate.
Movie S9. Time-lapse dark-field imaging of a six-input Disassembly gate.
Movie S10. Time-lapse dark-field imaging of a two-input Disassembly AND gate with three outputs.
Movie S11. Time-lapse dark-field imaging of a two-layer AND-AND cascade circuit.
Movie S12. Time-lapse dark-field imaging of a two-layer AND-OR cascade circuit.
Movie S13. Time-lapse dark-field imaging of a nanoparticle multiplexer circuit.
Reference (47)
REFERENCES AND NOTES
- 1.Prakash M., Gershenfeld N., Microfluidic bubble logic. Science 315, 832–835 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Nikitin M. P., Shipunova V. O., Deyev S. M., Nikitin P. I., Biocomputing based on particle disassembly. Nat. Nanotechnol. 9, 716–722 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Gines G., Zadorin A. S., Galas J.-C., Fujii T., Estevez-Torres A., Rondelez Y., Microscopic agents programmed by DNA circuits. Nat. Nanotechnol. 12, 351–359 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Yashin R., Rudchenko S., Stojanovic M. N., Networking particles over distance using oligonucleotide-based devices. J. Am. Chem. Soc. 129, 15581–15584 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Benenson Y., Biomolecular computing systems: Principles, progress and potential. Nat. Rev. Genet. 13, 455–468 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Katz E., Privman V., Enzyme-based logic systems for information processing. Chem. Soc. Rev. 39, 1835–1857 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Seelig G., Soloveichik D., Zhang D. Y., Winfree E., Enzyme-free nucleic acid logic circuits. Science 314, 1585–1588 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Qian L., Winfree E., Scaling up digital circuit computation with DNA strand displacement cascades. Science 332, 1196–1201 (2011). [DOI] [PubMed] [Google Scholar]
- 9.Green A. A., Kim J., Ma D., Silver P. A., Collins J. J., Yin P., Complex cellular logic computation using ribocomputing devices. Nature 548, 117–121 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chatterjee G., Dalchau N., Muscat R. A., Phillips A., Seelig G., A spatially localized architecture for fast and modular DNA computing. Nat. Nanotechnol. 12, 920–927 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Stojanovic M. N., Stefanovic D., A deoxyribozyme-based molecular automaton. Nat. Biotechnol. 21, 1069–1074 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Genot A. J., Bath J., Turberfield A. J., Reversible logic circuits made of DNA. J. Am. Chem. Soc. 133, 20080–20083 (2011). [DOI] [PubMed] [Google Scholar]
- 13.DelRosso N. V., Hews S., Spector L., Derr N. D., A molecular circuit regenerator to implement iterative strand displacement operations. Angew. Chem. Int. Ed. 56, 4443–4446 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Engelen W., Meijer L. H. H., Somers B., de Greef T. F. A., Merkx M., Antibody-controlled actuation of DNA-based molecular circuits. Nat. Commun. 8, 14473 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Silva A. P., Uchiyama S., Molecular logic and computing. Nat. Nanotechnol. 2, 399–410 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Kumar A., Kim S., Nam J.-M., Plasmonically engineered nanoprobes for biomedical applications. J. Am. Chem. Soc. 138, 14509–14525 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Wang H., Brandl D. W., Nordlander P., Halas N. J., Plasmonic nanostructures: Artificial molecules. Acc. Chem. Res. 40, 53–62 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Zhou B., Shi B., Jin D., Liu X., Controlling upconversion nanocrystals for emerging applications. Nat. Nanotechnol. 10, 924–936 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Urban A. S., Fedoruk M., Horton M. R., Rädler J. O., Stefani F. D., Feldmann J., Controlled nanometric phase transitions of phospholipid membranes by plasmonic heating of single gold nanoparticles. Nano Lett. 9, 2903–2908 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Liu L., Corma A., Metal catalysts for heterogeneous catalysis: From single atoms to nanoclusters and nanoparticles. Chem. Rev. 118, 4981–5079 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reddy L. H., Arias J. L., Nicolas J., Couvreur P., Magnetic nanoparticles: Design and characterization, toxicity and biocompatibility, pharmaceutical and biomedical applications. Chem. Rev. 112, 5818–5878 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Yan Y., Warren S. C., Fuller P., Grzybowski B. A., Chemoelectronic circuits based on metal nanoparticles. Nat. Nanotechnol. 11, 603–608 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Talapin D. V., Lee J.-S., Kovalenko M. V., Shevchenko E. V., Prospects of colloidal nanocrystals for electronic and optoelectronic applications. Chem. Rev. 110, 389–458 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Song T., Liang H., Synchronized assembly of gold nanoparticles driven by a dynamic DNA-fueled molecular machine. J. Am. Chem. Soc. 134, 10803–10806 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Liu D., Chen W., Sun K., Deng K., Zhang W., Wang Z., Jiang X., Resettable, multi-readout logic gates based on controllably reversible aggregation of gold nanoparticles. Angew. Chem. Int. Ed. 50, 4103–4107 (2011). [DOI] [PubMed] [Google Scholar]
- 26.Motornov M., Zhou J., Pita M., Gopishetty V., Tokarev I., Katz E., Minko S., “Chemical Transformers” from nanoparticle ensembles operated with logic. Nano Lett. 8, 2993–2997 (2008). [DOI] [PubMed] [Google Scholar]
- 27.von Maltzahn G., Harris T. J., Park J.-H., Min D.-H., Schmidt A. J., Sailor M. J., Bhatia S. N., Nanoparticle self-assembly gated by logical proteolytic triggers. J. Am. Chem. Soc. 129, 6064–6065 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu J., Lu Y., Smart nanomaterials responsive to multiple chemical stimuli with controllable cooperativity. Adv. Mater. 18, 1667–1671 (2006). [Google Scholar]
- 29.Angelos S., Yang Y.-W., Khashab N. M., Stoddart J. F., Zink J. I., Dual-controlled nanoparticles exhibiting AND logic. J. Am. Chem. Soc. 131, 11344–11346 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Freeman R., Finder T., Willner I., Multiplexed analysis of Hg2+ and Ag+ ions by nucleic acid functionalized CdSe/ZnS quantum dots and their use for logic gate operations. Angew. Chem. Int. Ed. 121, 7958–7961 (2009). [DOI] [PubMed] [Google Scholar]
- 31.Bray D., Protein molecules as computational elements in living cells. Nature 376, 307–312 (1995). [DOI] [PubMed] [Google Scholar]
- 32.Bray D., Intracellular signalling as a parallel distributed process. J. Theor. Biol. 143, 215–231 (1990). [DOI] [PubMed] [Google Scholar]
- 33.Antebi Y. E., Linton J. M., Klumpe H., Bintu B., Gong M., Su C., McCardell R., Elowitz M. B., Combinatorial signal perception in the BMP pathway. Cell 170, 1184–1196.e24 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salaita K., Nair P. M., Petit R. S., Neve R. M., Das D., Gray J. W., Groves J. T., Restriction of receptor movement alters cellular response: Physical force sensing by EphA2. Science 327, 1380–1385 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang D. Y., Seelig G., Dynamic DNA nanotechnology using strand-displacement reactions. Nat. Chem. 3, 103–113 (2011). [DOI] [PubMed] [Google Scholar]
- 36.Kim S., Park J.-E., Hwang W., Seo J., Lee Y.-K., Hwang J.-H., Nam J.-M., Optokinetically encoded nanoprobe-based multiplexing strategy for microRNA profiling. J. Am. Chem. Soc. 139, 3558–3566 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Klajn R., Stoddart J. F., Grzybowski B. A., Nanoparticles functionalised with reversible molecular and supramolecular switches. Chem. Soc. Rev. 39, 2203–2237 (2010). [DOI] [PubMed] [Google Scholar]
- 38.Langecker M., Arnaut V., List J., Simmel F. C., DNA nanostructures interacting with lipid bilayer membranes. Acc. Chem. Res. 47, 1807–1815 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Ohta S., Glancy D., Chan W. C. W., DNA-controlled dynamic colloidal nanoparticle systems for mediating cellular interaction. Science 351, 841–845 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Jung C., Allen P. B., Ellington A. D., A stochastic DNA walker that traverses a microparticle surface. Nat. Nanotechnol. 11, 157–163 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adamala K. P., Martin-Alarcon D. A., Guthrie-Honea K. R., Boyden E. S., Engineering genetic circuit interactions within and between synthetic minimal cells. Nat. Chem. 9, 431–439 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Regot S., Macia J., Conde N., Furukawa K., Kjellén J., Peeters T., Hohmann S., de Nadal E., Posas F., Solé R., Distributed biological computation with multicellular engineered networks. Nature 469, 207–211 (2010). [DOI] [PubMed] [Google Scholar]
- 43.Ba H., Rodríguez-Fernández J., Stefani F. D., Feldmann J., Immobilization of gold nanoparticles on living cell membranes upon controlled lipid binding. Nano Lett. 10, 3006–3012 (2010). [DOI] [PubMed] [Google Scholar]
- 44.Zhang D. Y., Turberfield A. J., Yurke B., Winfree E., Engineering entropy-driven reactions and networks catalyzed by DNA. Science 318, 1121–1125 (2007). [DOI] [PubMed] [Google Scholar]
- 45.D. Y. Zhang, in DNA Computing and Molecular Programming, Y. Sakakibara, Y. Mi, Eds. (Springer Berlin Heidelberg, 2011), pp. 162–175. [Google Scholar]
- 46.Zadeh J. N., Steenberg C. D., Bois J. S., Wolfe B. R., Pierce M. B., Khan A. R., Dirks R. M., Pierce N. A., NUPACK: Analysis and design of nucleic acid systems. J. Comput. Chem. 32, 170–173 (2011). [DOI] [PubMed] [Google Scholar]
- 47.Lee Y. K., Kim S., Oh J.-W., Nam J.-M., Massively parallel and highly quantitative single-particle analysis on interactions between nanoparticles on supported lipid bilayer. J. Am. Chem. Soc. 136, 4081–4088 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/2/eaau2124/DC1
Fig. S1. Conceptual illustrations of the LNT platform.
Fig. S2. Characterization of nanoparticles.
Fig. S3. Diffusion of nanoparticles tethered to an SLB.
Fig. S4. Design and implementation of a Disassembly YES gate.
Fig. S5. Tethering of nanoparticles to an SLB.
Fig. S6. Image analysis pipeline.
Fig. S7. Scattering signal profiles of red, green, and blue nanoparticles.
Fig. S8. Nucleotide-level schematics of two-input logic gates.
Fig. S9. Design principles for nanoparticle logic gates.
Fig. S10. Modularity of a hairpin-based two-input Assembly AND gate.
Fig. S11. Uneven responses of a two-input Disassembly OR gate.
Fig. S12. A dual-rail NAND gate.
Fig. S13. Disassembly INHIBIT and six-input Disassembly gates.
Fig. S14. Complex multi-input Disassembly gates.
Fig. S15. Dark-field snapshots of two-layer AND-AND and AND-OR cascade circuits.
Fig. S16. Two-layer OR-AND and INHIBIT-AND cascade circuits.
Fig. S17. A two-layer OR-OR cascade circuit.
Fig. S18. A nanoparticle multiplexer circuit.
Fig. S19. Effect of DNA concentration on floater diffusion.
Table S1. Response rates of two-input logic gates.
Table S2. DNA sequences of thiolated strands used for functionalizing nanoparticles.
Table S3. DNA sequences and experimental conditions used in circuit operations.
Movie S1. Time-lapse dark-field imaging of a nanoparticle Assembly YES gate.
Movie S2. Time-lapse dark-field imaging of a nanoparticle Disassembly YES gate.
Movie S3. Receptor-only visualization of a dark-field movie.
Movie S4. Time-lapse dark-field imaging of a two-input Assembly AND gate.
Movie S5. Time-lapse dark-field imaging of a two-input Assembly OR gate.
Movie S6. Time-lapse dark-field imaging of a two-input Disassembly AND gate.
Movie S7. Time-lapse dark-field imaging of a two-input Disassembly OR gate.
Movie S8. Time-lapse dark-field imaging of a two-input Disassembly INHIBIT gate.
Movie S9. Time-lapse dark-field imaging of a six-input Disassembly gate.
Movie S10. Time-lapse dark-field imaging of a two-input Disassembly AND gate with three outputs.
Movie S11. Time-lapse dark-field imaging of a two-layer AND-AND cascade circuit.
Movie S12. Time-lapse dark-field imaging of a two-layer AND-OR cascade circuit.
Movie S13. Time-lapse dark-field imaging of a nanoparticle multiplexer circuit.
Reference (47)


