Figure 6.
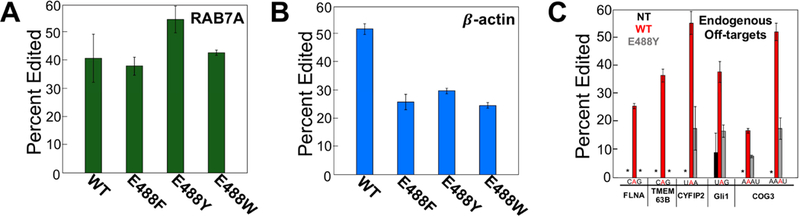
Directed editing on endogenous targets and observed endogenous off-targets. (A) Directed editing on endogenous 3’-UTR of RAB7A with overexpression of hADAR2 and cytidine guide RNA or overexpression of bulky mutants hADAR2 E488X (X = F, Y, W) and reduced abasic guide RNA. (B) Directed editing on endogenous 3’-UTR of β-actin with overexpression of hADAR2 and cytidine guide RNA or overexpression of bulky mutants hADAR2 E488X (X = F, Y, W) and reduced abasic guide RNA. (C) Percent editing on various endogenous off-target transcripts: FLNA, TMEM63B, CYFIP2, Gli1 and COG3. Bolded in red is off-target site with 5’ and 3’ flanking sequence. Bar shown in black is percent editing of off-target with no transfection. Bars shown in red are percent editing of off-targets when directing editing at endogenous RAB7A with overexpression of wildtype hADAR2 and cytidine guide RNA. Bars shown in grey are percent editing of off-targets when directing editing at endogenous RAB7A with overexpression of hADAR2 E488Y and reduced abasic guide RNA. Percent editing values can be found in Table S5. An asterisk (*) indicates no detected editing. Error bars, s.d. (n = 3 biological replicates).
