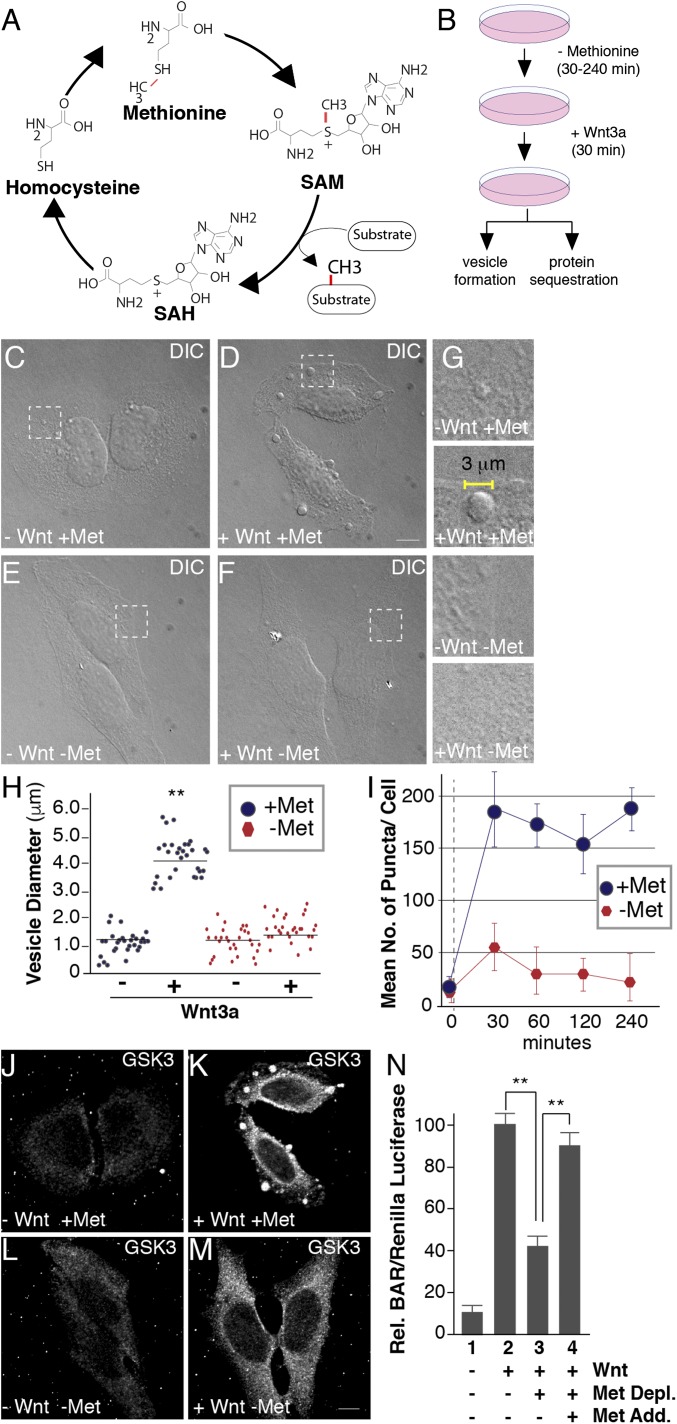
Fig. 1.
Methionine depletion inhibits Wnt signaling. (A) The universal cofactor for methylation SAM is generated through a biochemical mechanism known as the methionine cycle. Methionine is converted into SAM through an ATP-driven reaction. Methyltransferases use SAM as a cofactor to transfer methyl groups (in red) to a myriad of substrates, generating the byproduct SAH. SAH is rapidly converted to homocysteine, which can become remethylated to form methionine, completing the cycle. (B) Experimental design used for testing the effect of methionine deprivation on Wnt-induced endocytosis. Cultured cells were preincubated in methionine-depleted medium for 30 min and then treated with Wnt3a for 30 min (in the case of HeLa cell phase microscopy analyses or in situ protease protection assays) or 12 h (for HEK293T BAR/β-catenin luciferase assays). (C–F) Treatment with Wnt3a for 30 min triggered the formation of large puncta visualized by phase microscopy in HeLa cells; methionine depletion for 30 min strikingly inhibited vesicle formation in the presence of Wnt3a (compare D to F); the indicated stippled rectangles were enlarged. (G) Enlargements of Wnt3a-induced vesicles (2–5 μm in diameter) which were absent minus methionine. (H) Quantification of the diameters of Wnt-induced vesicles using ImageJ. (I) Time-course analysis of methionine depletion for 30, 60, 120, or 240 min followed by 30 min of Wnt3a signaling; note the significant decrease in vesicle formation after only 30 min (despite using nondialyzed serum in this case). (J–M) Wnt-induced GSK3 sequestration inside vesicles was inhibited by methionine starvation for 30 min, as assessed by in situ proteinase K protection assay (compare K to M). (N) Methionine depletion inhibited Wnt signaling in BAR luciferase reporter assays, while methionine readdition in column 4, rescued Wnt inhibition. Wnt luciferase reporter (BAR) was normalized using Renilla (49). (Scale bars: 10 μm.) **P < 0.01; n < 3.