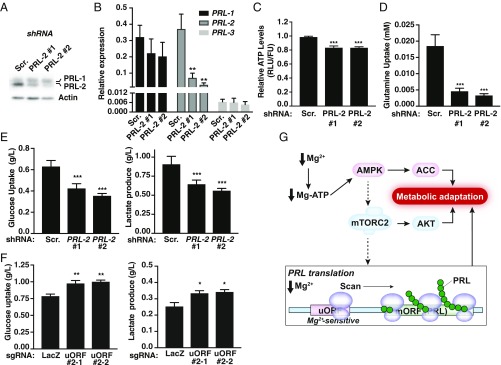
Fig. 6.
PRL-2 mediates cellular metabolic reprogramming. (A) Western blot analysis of MDA-MB-231 cells expressing dox-inducible PRL-2 or scramble (Scr.) shRNAs treated with dox for 48 h. (B) Quantification of PRL-1, -2, and -3 mRNAs was performed on dox-inducible PRL-2 or Scr. shRNA-expressing cells treated with dox for 48 h. Data are mean ± SD (n = 3); **P < 0.01 vs. Scr. by two-way ANOVA. (C–E) MDA-MB-231 cells expressing dox-inducible PRL-2 or Scr. shRNAs were treated with dox for 48 h and the indicated metabolites measured. All plots are representative of at least three independent experiments. Data are means ± SD (n = 4–5); ***P < 0.001 vs. Scr. by two-way ANOVA. (F) Metabolite measurement of MDA-MB-231 cells expressing the two sgRNAs targeting the uORF cultured in 2.5 mM glucose. All plots are representative of at least three independent experiments. Data are means ± SD (n = 4); **P < 0.01 or *P < 0.05 vs. LacZ by two-way ANOVA. (G) Schematic depicting how low intracellular magnesium leads to an increase in PRL protein levels. When magnesium is reduced, the AMPK/mTORC2-dependent pathways, which are generally involved in regulating cellular bioenergetics, get activated to promote a rise in PRL protein synthesis to contribute to the observed metabolic switch. Concomitantly, a uORF contributes to the regulation of PRL mRNA translation by a “magnesium-sensing” mechanism involving scanning ribosomes that will be stalled at the uORF in the presence of magnesium but become competent for reinitiation at the downstream start codon of the main ORF (mORF) under conditions of low magnesium, leading to PRL protein synthesis.