Fig. 6.
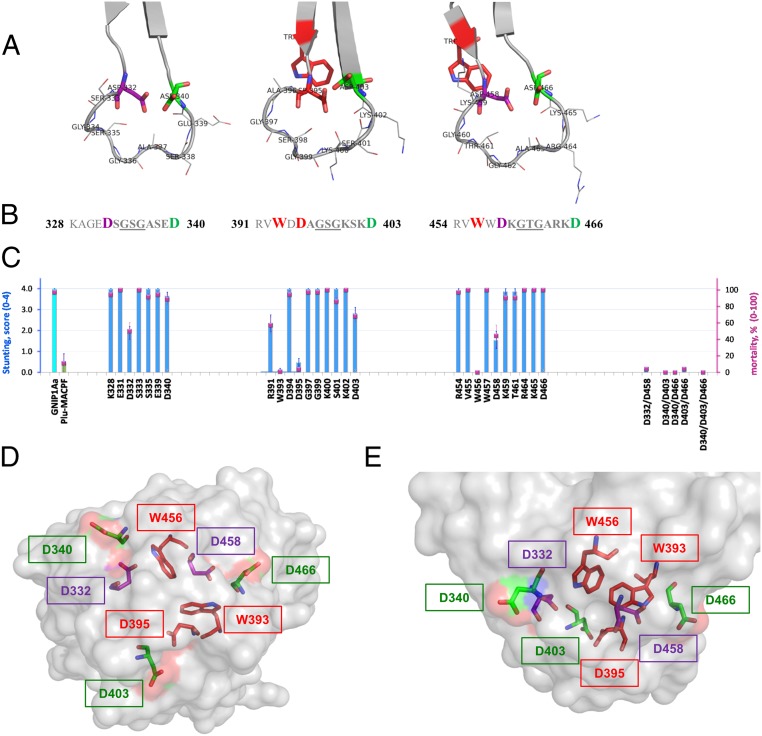
Structure–function analysis of three conserved loops of the C-terminal domain of GNIP1Aa. (A) Ribbon representation of three apical loops. Functionally important and conserved residues (Asp and Trp) are framing each loop, shown in stick representation and labeled with a corresponding residue number. The loop-composing residues are labeled and shown in lines representation. (B) Sequences of the three loop regions with the flanking regions, colored the same as in A. Red bold: residues W393 (#−2 in consensus, Fig. 3), D395 (#1), and W456 (#−2) are unique; replacement of any one completely abolishes protein toxicity. Purple bold: Asp residues in position #1 of the first and third loops, D332 and D458, are essential for GNIP1Aa activity. Green bold: Asp residues in position #9, D340, D403, and D466, for all three loops are fairly important. (C) Activity of the selected single-site Ala mutants (from the left: three groups of data), double and triple mutants (on the right side) in WCR bio-assays, shown in stunt (blue bars) and mortality (magenta squares) values. See more details in SI Appendix, Fig. S3. The GNIP1Aa is the wild-type GNIP1Aa, the positive control; its stunting activity is represented by a cyan-color bar. Plu-MACPF is the negative control with low background WCR activity, a green-colored bar for stunt. The letter and number indicate a one-letter code and position number for a residue within the wild-type GNIP1Aa1 protein, replaced by an alanine. (D) Semitransparent space-filling representation of the C-terminal domain surface with the conserved residues in positions #−2 (Trp), #1 (Asp), and #9 (Asp) colored the same as in A and B and shown in stick representation. A view along the threefold pseudosymmetry axis from the apical tip of the domain, opposite side of the view shown in Fig. 2C. (E) The side view on the apical tip of the C-terminal domain rotated 90° relative to D. The same view as in Fig. 2D.