Significance
Group VII ethylene response factors (ERFVIIs) function as oxygen sensors via the N-end rule pathway of proteolysis. SUB1A-1, an ERFVII, is a “master regulator” of submergence tolerance in rice, but escapes the N-end rule pathway despite containing the canonical N-degron. This raises questions about how rice senses hypoxia stress during submergence. Here, two ERFVIIs, ERF66 and ERF67, are identified as direct transcriptional targets of SUB1A-1 that are substrates of the N-end rule pathway and promote survival of submergence. We propose a regulatory cascade involving SUB1A-1 and ERF66/ERF67 as a response to submergence stress in rice. Furthermore, the SUB1A-1 C terminus interacts with the SUB1A-1 N terminus and prevents its turnover, which may explain how SUB1A-1 evades N-end rule pathway.
Keywords: submergence, rice, ethylene response factors, transcriptional regulation, N-end rule pathway
Abstract
The rice SUB1A-1 gene, which encodes a group VII ethylene response factor (ERFVII), plays a pivotal role in rice survival under flooding stress, as well as other abiotic stresses. In Arabidopsis, five ERFVII factors play roles in regulating hypoxic responses. A characteristic feature of Arabidopsis ERFVIIs is a destabilizing N terminus, which functions as an N-degron that targets them for degradation via the oxygen-dependent N-end rule pathway of proteolysis, but permits their stabilization during hypoxia for hypoxia-responsive signaling. Despite having the canonical N-degron sequence, SUB1A-1 is not under N-end rule regulation, suggesting a distinct hypoxia signaling pathway in rice during submergence. Herein we show that two other rice ERFVIIs gene, ERF66 and ERF67, are directly transcriptionally up-regulated by SUB1A-1 under submergence. In contrast to SUB1A-1, ERF66 and ERF67 are substrates of the N-end rule pathway that are stabilized under hypoxia and may be responsible for triggering a stronger transcriptional response to promote submergence survival. In support of this, overexpression of ERF66 or ERF67 leads to activation of anaerobic survival genes and enhanced submergence tolerance. Furthermore, by using structural and protein-interaction analyses, we show that the C terminus of SUB1A-1 prevents its degradation via the N-end rule and directly interacts with the SUB1A-1 N terminus, which may explain the enhanced stability of SUB1A-1 despite bearing an N-degron sequence. In summary, our results suggest that SUB1A-1, ERF66, and ERF67 form a regulatory cascade involving transcriptional and N-end rule control, which allows rice to distinguish flooding from other SUB1A-1–regulated stresses.
Floods are climate-related catastrophes that severely influence plant growth, survival, and reproduction. Flooding stress includes waterlogging, when only roots are exposed to soil flooded with water, and submergence, when the shoots are partially or completely immersed in water (1). Under flooding stress, oxygen deprivation prevents aerobic respiration and limits ATP synthesis, resulting in a severe energy crisis (2). The alternative energy supply from NAD+ regeneration using anaerobic fermentation is not a sufficient strategy, as it accumulates toxic metabolites (3).
Two opposite growth-related flooding survival strategies have evolved in rice: escape and quiescence. The escape strategy is transcriptionally regulated in certain deepwater cultivars by the group VII ethylene response factors (ERFVIIs) SNORKEL1 and 2 and in other varieties through control of gibberellin production by the transcription factor OsEIL1 (4–6). In each of these cases, the rice plant adapts to flooding by promoting internode elongation to grow above the water level, which allows gas exchange with the atmosphere and thereby prevents the onset of hypoxia in cells. For the quiescence strategy, a few rice cultivars, such as FR13A, show high tolerance and survive as long as 2 wk under complete submergence as a result of the presence of the SUBMERGENCE 1 (Sub1) locus, which consists of a cluster of three Oryza sativa ERFVIIs (OsERFVIIs) that are related to SNORKEL1/2 but function differently (5). Among them, SUB1A-1 functions as a “master regulator,” coordinating the quiescence responses required for survival of prolonged submergence (5). Submergence-intolerant cultivars, such as Swarna and IR64, lack SUB1A-1 or have the SUB1A-2 allele, which is inactive as a result of a point mutation within the coding region (5, 7). Introgression or overexpression of SUB1A-1 into the Swarna and IR64 lines confers significant submergence tolerance (5, 8, 9).
In Arabidopsis, five ERFVIIs, including HYPOXIA RESPONSIVE ERF (HRE) 1, HRE2, RELATED TO APETALA (RAP) 2.2, RAP2.3, and RAP2.12 (10), play some roles in regulating hypoxic responses. Overexpressing individual Arabidopsis thaliana ERFVIIs (AtERFVIIs) improves tolerance to hypoxic or flooding stress. Conversely, KO or knockdown lines of AtERFVII genes are more susceptible to flooding stress (11–17). It is proposed that each ERFVII likely has distinct and overlapping targets that orchestrate expression of hypoxia response genes in Arabidopsis (18). One characteristic feature of AtERFVIIs is a highly conserved N terminus that starts with the MCGGAI(I/L) motif. In vitro and in vivo analyses of protein stability showed that this conserved motif functions as an N-degron, which promotes the degradation of ERFVIIs via the oxygen- and NO-dependent N-end rule pathway of targeted proteolysis (19–23). In this pathway, methionine aminopeptidase (MetAP) first removes the methionine residue from the N-terminal Met-Cys, leaving cysteine as the first residue. Under normoxia, the N-terminal Cys residue is subjected to oxygen-dependent oxidation by plant cysteine oxidases, which convert Cys to negatively charged Cys-sulfinic acid (CysO2) (24, 25). The N-terminal CysO2 is then arginylated by arginyl tRNA transferase 1. Finally, ERFVIIs with N-terminal Arg-CysO2 are proposed to be recognized by the N-recognin E3 ligase proteolysis 6 (PRT6) and degraded via the ubiquitin–proteasome pathway. Under hypoxia, Cys oxidation is limited, which subsequently prevents degradation via the N-end rule pathway, so the AtERFVIIs are stabilized and accumulate to transcriptionally trigger downstream hypoxic responses.
In contrast to Arabidopsis, the rice genome consists of 18 ERFVIIs, some of which are cultivar-specific, such as SUB1A-1 and SNORKEL1/2. SUB1A-1 and SNORKEL1/2 play key regulatory roles in FR13A and deepwater rice, respectively, in response to flooding stress (4, 5, 10). The involvement of AtERFVIIs and SUB1A-1 in activating hypoxic responsive and fermentative genes during submergence suggests that they have similar functions as master regulators of hypoxic responses in Arabidopsis and rice, respectively. However, ectopic expression of SUB1A-1 in Arabidopsis cannot enhance tolerance to submergence in the dark (26). Despite possessing a similar Met-Cys–initiating N-terminal degron sequence as the AtERFVIIs, SUB1A-1 is not subject to regulation by the N-end rule pathway in vitro (19). The ability of SUB1A-1 to escape degradation through the N-end rule pathway may be key to its involvement in other abiotic stress responses, such as surviving reactive oxygen species accumulation and rapid dehydration following desubmergence, and prolonged darkness (27, 28). It is generally believed that SUB1A-1 may serve a key signaling hub that regulates responses to various stresses independently of oxygen levels. This raises two critical questions as to (i) how oxygen sensing is regulated in rice and (ii) how SUB1A-1 escapes N-end rule regulation.
Herein, we report that two rice ERFVIIs, ERF66 and ERF67, function downstream of SUB1A-1 to form a regulatory cascade in response to submergence stress. ERF66 and ERF67 are induced under submergence in a SUB1A-1–dependent manner and are direct transcriptional targets of SUB1A-1. In contrast to SUB1A-1, ERF66 and ERF67 are subjected to oxygen-dependent turnover via the N-end rule pathway. Overexpression of GST-tagged ERF66/67 in the submergence-sensitive Tainung 67 (TNG67) cultivar resulted in enhanced expression of genes associated with submergence tolerance and increased submergence survival. NMR structural analysis of the SUB1A-1 N terminus revealed a flexible, random coil structure that should permit interaction with N-end rule enzymatic components and therefore degradation. However, we found that the C-terminal region of SUB1A-1 prevents its degradation and directly interacts with the SUB1A-1 N terminus, providing insight into how SUB1A-1 evades degradation under hypoxia. We propose that the flooding response in SUB1A-1–encoding cultivars involves SUB1A-1–dependent transcriptional activation of ERF66 and ERF67, which are then stabilized only under hypoxia to coordinate the submergence-specific response, thereby allowing rice plants to discriminate flooding from other SUB1A-1–regulated stresses.
Results
SUB1A-1 Regulates ERFVII Gene Expression During Submergence.
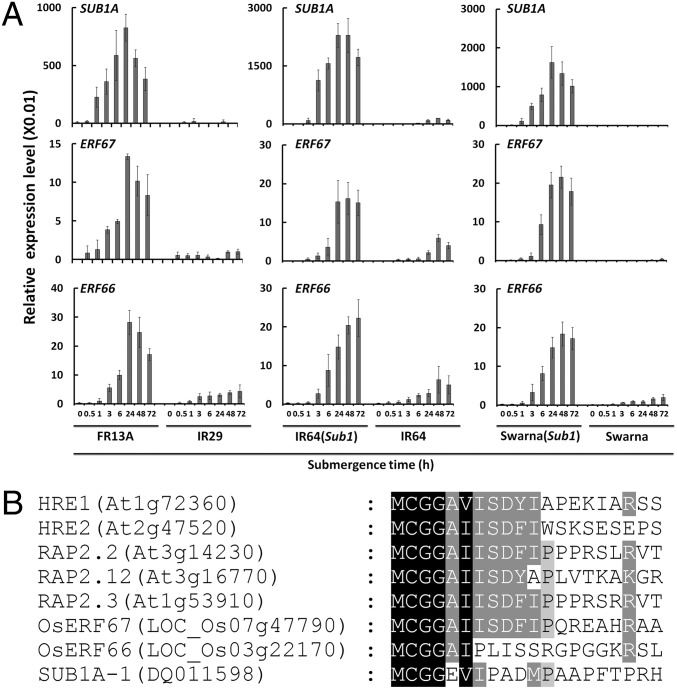
To understand the transcriptional networks regulated by SUB1A-1 during submergence, we dissected the transcriptional profiles of 16 OsERFVIIs (all except SNORKEL1 and 2, which are absent in most cultivars) in two Indica rice cultivars that display contrasting sensitivity toward submergence stress. FR13A, the submergence-tolerant cultivar, possesses the tolerant SUB1A-1 allele. IR29, the submergence-sensitive cultivar, possesses the intolerant SUB1A-2 allele, which contains an inactive SUB1A-2 as a result of a single amino acid substitution at position 186 from serine (SUB1A-1) to proline (SUB1A-2) (5, 7). By comparing the results of quantitative RT-PCR (qRT-PCR; SI Appendix, Fig. S1), we found that the transcript levels of ERF59, ERF60, ERF61, ERF66, and ERF67 were much higher under submergence in FR13A than in IR29. It is reported that ERF73/SUB1C is negatively regulated by SUB1A-1 (5, 8). Consistently, our result shows that the transcript level of ERF73/SUB1C was much lower in FR13A than in IR29 (SI Appendix, Fig. S1). To eliminate the difference in transcriptional levels that arise from different genetic backgrounds, we then compared OsERFVII expression profiles of submergence-sensitive cultivars, IR64 and Swarna, with those of corresponding near-isogenic lines with introgressed SUB1A-1. The results showed that only the transcripts of ERF66 and ERF67 were significantly more abundant in the SUB1A-1 introgressed cultivars IR64(Sub1) and Swarna(Sub1) than in submergence-sensitive IR64 and Swarna (Fig. 1A and SI Appendix, Figs. S2 and S3). Jung et al. (29) previously found that ERF66, ERF67, and ERF68 were induced in the Sub1 near-isogenic line of M202 by using microarray approaches. Our results also showed that ERF68 responds to submergence within 30 min, but we found no differences in ERF68 expression between WT cultivars and corresponding SUB1A-1 lines (SI Appendix, Figs. S1–S3). In contrast, our data showed that ERF66 and ERF67 are up-regulated only in the presence of SUB1A-1 upon submergence (Fig. 1A).
Fig. 1.
Sub1A, ERF66, and ERF67 show similar transcriptional expression patterns during submergence. (A) Transcriptional profiling of SUB1A-1, ERF66, and ERF67 under submergence in FR13A, IR 29, IR64(Sub1), IR64, Swarna(Sub1), and Swarna. Fourteen-day-old seedlings were subjected to submergence treatment, and aerial tissues were harvested at the indicated time points. Transcript levels of Sub1A, ERF66, and ERF67 were quantified by qRT-PCR. Relative expression level is determined by ΔCT of Sub1A, ERF66, and ERF67 normalized by tubulin mRNA level as the internal control. The data represent means ± SD from three independent replicates. (B) N-terminal amino acid sequence alignment of five AtERFVIIs, ERF66, ERF67, and SUB1A-1.
SUB1A-1 Directly Activates ERF66 and ERF67.
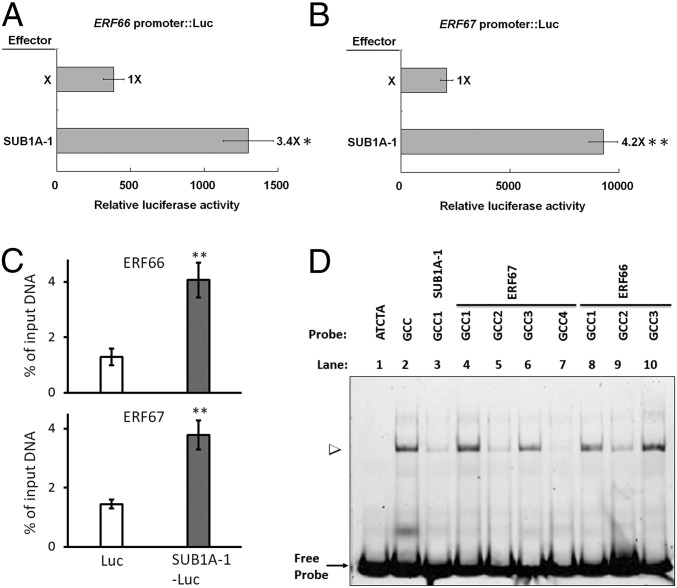
Next, we used protoplast transient assays to examine whether SUB1A-1 directly controls the transcription of ERF66 and ERF67 genes. By using an effector construct encoding SUB1A-1 driven by the Ubiquitin promoter (UbiP), cotransformed with a reporter construct encoding luciferase (Luc) driven by the ERF66 promoter (SI Appendix, Fig. S4A), we found a three- to fourfold increase in Luc activity compared with the control (Fig. 2A). Similarly, SUB1A-1 stimulated transcription from the ERF67 promoter four- to fivefold (Fig. 2B). These results show that SUB1A-1 transcriptionally activates the ERF66 and ERF67 genes. We then confirmed the direct binding of SUB1A-1 with the ERF66/67 promoter region by ChIP/quantitative PCR (qPCR; Fig. 2C and SI Appendix, Fig. S4B), observing a two- to threefold enrichment of ERF66 and ERF67 promoter sequences in SUB1A-1 immunoprecipitate compared with the control.
Fig. 2.
SUB1A-1 transactivates the ERF66 and ERF67 promoters through interacting with GCC boxes. (A and B) Transactivation assay in rice protoplasts showing SUB1A-1–dependent activation of ERF66 and ERF67 promoters, respectively. The SUB1A-1 coding region was linked to UbiP for use as an effector construct (SI Appendix, Fig. S4A). The promoter sequences of ERF66 and ERF67 were fused to the coding sequence of Luc to be used as reporter constructs (SI Appendix, Fig. S4A). A UbiP::GUS plasmid was used as an internal control. Relative Luc activity of effector genes (calculated as the ratio of Luc activity/GUS activity/total proteins in micrograms) was then compared with the control. (C) SUB1A-1-Luc–specific enrichment of ERF66/67 promoter sequences using ChIP-qPCR. The data represent mean ± SD from three replicates (*P < 0.05 and **P < 0.01 indicate significant differences by Student’s t test). (D) EMSA assays of the interaction between recombinant SUB1A-1 and FAM-labeled DNA. Each GCC contains a different flanking sequence (SI Appendix, Fig. S4B and Table S3). In the control experiment (lanes 1 and 2), SUB1A-1 has no binding with reference ATCTA probe (59) and binds to reference GCC DNA (60). The ERF67-GCC1 and ERF66-GCC1 show similar binding affinity to SUB1A-1, and SUB1A-1-GCC1, ERF67-GCC2, ERF67-GCC4, and ERF66-GCC2 show much weaker binding affinity to SUB1A-1, compared with the reference GCC.
The conserved APETALA2 (AP2) domain of ERFVIIs is known to interact with a GCC box with a core sequence GCCGCC (30–32). Multiple-GCC boxes with various flanking sequences are found in ERF66 and ERF67 promoters, and one is found in SUB1A-1 promoter (SI Appendix, Fig. S4B). Our EMSA assays with recombinant SUB1A-1 show that SUB1A-1 preferably interacts with ERF66-GCC1, ERF66-GCC3, ERF67-GCC1, and ERF67-GCC3, but not SUB1A-1-GCC1 or other GCC boxes in the promoters of ERF66 or ERF67 (Fig. 2D). To eliminate the possibility that the fluorescent probe may interfere with the binding, we confirmed our findings with competition assays by using unlabeled GCC boxes (SI Appendix, Fig. S4C). In a more recent study, RAP2.2 and RAP2.12 could bind to an extended GCC consensus sequence, designated the Arabidopsis hypoxia-responsive promoter element, to activate core hypoxia response genes in Arabidopsis (33). Our EMSA results extend the current knowledge by demonstrating that the flanking sequence of the GCC boxes in the promoters of ERF66 and ERF67 is also important and may play roles in SUB1A-1 selectivity for transcriptional activation.
Together, our experiments (Figs. 1 and 2) show that SUB1A-1 directly up-regulates ERF66 and ERF67 in response to submergence through interacting with GCC boxes in their respective promoter regions.
Overexpression of ERF66, ERF67, or SUB1A-1 Enhances Submergence Tolerance in Transgenic Rice.
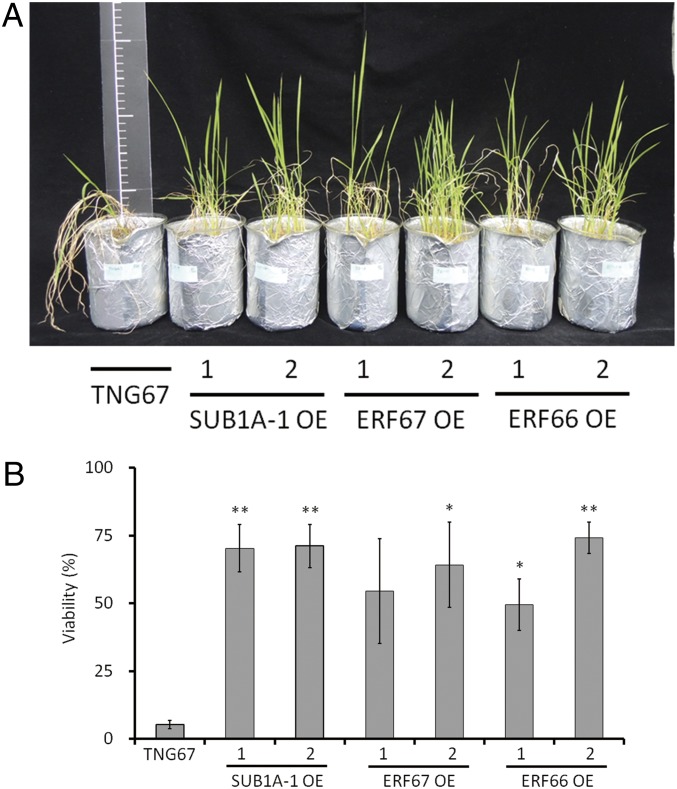
As ERF66 and ERF67 are downstream targets of SUB1A-1, we next examined if ERF66 and ERF67 participate in submergence tolerance. We individually overexpressed each gene (SI Appendix, Fig. S5) in the submergence-sensitive TNG67 cultivar (which does not contain SUB1A-1) and assessed viability of the resulting transgenic lines following 7 d of submergence. By using two independent lines for each transgene, we found that all three transcription factors, including SUB1A-1, individually led to enhanced submergence tolerance compared with WT (Fig. 3). In contrast to Xu et al. (5), we did not observe a semidwarf phenotype in the TNG67 SUB1A-1 overexpressing (SUB1A-1 OE) lines, which may be related to the use of different genetic backgrounds in the two studies.
Fig. 3.
Phenotypes of SUB1A-1, ERF67, and ERF66 overexpression lines after submergence. (A) Rice plants after 14 d of recovery from submergence. Fourteen-day-old rice plants were submerged for 7 d in darkness. After submergence, plants were returned to normal growth conditions for 14 d of recovery and photographed. (B) Viability of whole plants after desubmergence. The whole plant viability of each genotype was evaluated in the samples shown in A. Plants were scored as viable if a new leaf appeared during the recovery period. The data represent means ± SD from two independent replicates (*P < 0.05 and **P < 0.01 indicate significant differences by Student’s t test). The P value of ERF67 OE line 1 is 0.06.
ERF66 and ERF67 Are Subject to N-End Rule Regulation.
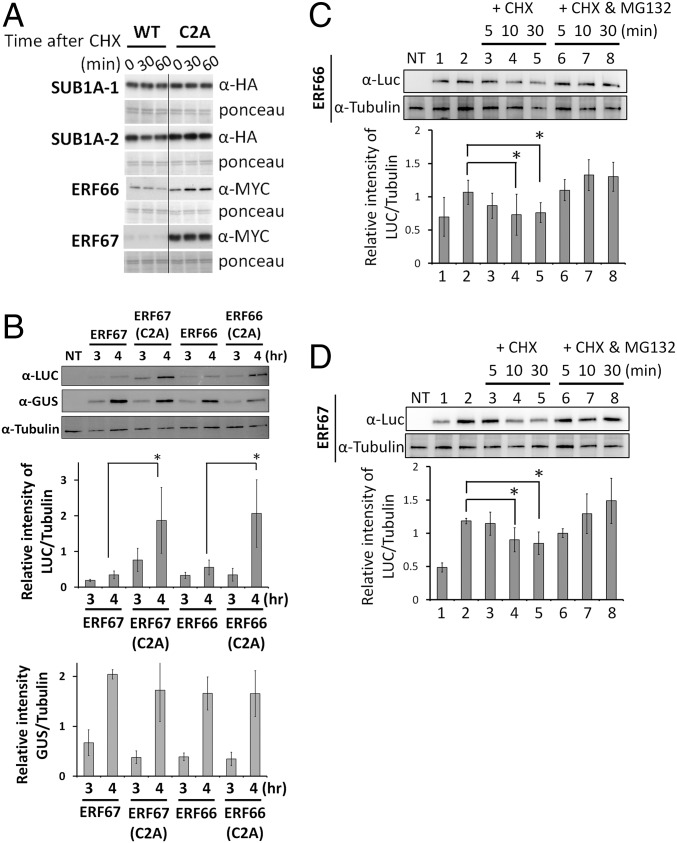
SUB1A-1, ERF66, and ERF67 all have the conserved N-degron sequence of MCGG (Fig. 1B and SI Appendix, Fig. S6), but SUB1A-1 is not subjected to N-end rule regulation in vitro (19). We next investigated if ERF66 and ERF67 are targets of the N-end rule pathway. By using a previously established in vitro assay (19), whereby proteins are expressed in a rabbit reticulocyte system containing conserved N-end rule components, we showed that a cysteine-to-alanine mutation at residue position 2 (C2A) in ERF66-MYC and ERF67-MYC led to enhanced protein stability in the presence of the protein synthesis inhibitor cycloheximide (CHX) compared with WT, whereas WT and C2A variants of SUB1A-1-HA showed no difference in protein stability (Fig. 4A). As N-terminal cysteine is crucial for turnover of Arabidopsis ERFVIIs (19, 20), these in vitro data suggest that ERF66 and ERF67, in contrast to SUB1A-1, are substrates of the N-end rule pathway.
Fig. 4.
ERF66 and ERF67 are substrates of the N-end rule pathway. (A) In vitro analysis of protein stability of HA- or MYC-tagged WT and C2A variants of SUB1A-1, SUB1A-2, ERF66, and ERF67 following treatment with CHX. (B) The stability of ERF66 and ERF67 expressed in rice protoplasts is enhanced by a C2A mutation or treatment with MG132. UbiP::ERFVII-Luc constructs were cotransfected into TNG67 rice protoplasts with a UbiP::GUS plasmid, which was used as a stable control. The transfected protoplasts were incubated in W5 solution for 3 or 4 h and then harvested for Western blot analysis (SI Appendix, Fig. S7A), and the relative levels of Luc and GUS were normalized to tubulin, respectively. NT, nontransfected. The molecular weights of ERF66/67-Luc fusion proteins are ∼87/85 kDa, respectively. (C and D) CHX chase of ERF66 and ERF67 with/without MG132 in rice protoplasts. The protein levels of ERF66/ERF67 4 h after transformation without MG132 (lane 1) and with MG132 (lane 2) are shown. CHX chase experiments were initiated after 4 h treatment with MG132 to ensure high levels of protein at the beginning of the chase by replacing buffer with CHX only (100 μM) or CHX and MG132 (20 μM). The relative levels of Luc were normalized to tubulin. The data represent means ± SD from three independent replicates (*P < 0.05 indicates a significant difference by Student’s t test).
We next examined the regulation of ERF66, ERF67, and SUB1A-1 stability in vivo by transiently expressing C-terminally Luc-tagged WT and C2A mutant variants driven by the Ubi promoter in TNG67 rice protoplast cells (Fig. 4B and SI Appendix, Fig. S7 A and B). Here we could detect only low levels of WT ERF66/67 by Western blot, but a C2A mutation or treatment with MG132 led to enhanced accumulation (Fig. 4 B–D). In contrast, WT and C2A variants of SUB1A-1 showed similar levels of accumulation (SI Appendix, Fig. S7C). Following treatment with CHX, the accumulated WT ERF66/67-Luc proteins were degraded over time but showed enhanced stability in the presence of MG132, confirming that observed differences in ERF66/67 levels are linked to their regulation via proteasome degradation pathway (Fig. 4 C and D). We also examined the influence of submergence-induced hypoxia on ERF66 and ERF67 stability by using a transgenic Arabidopsis approach. Here we observed increased protein levels of GFP-tagged ERF66 and ERF67 during a submergence time course (SI Appendix, Fig. S8A), whereas transcript change remained relatively constant compared with the hypoxia-inducible control gene ALCOHOL DEHYDROGENASE1 (ADH1; SI Appendix, Fig. S8B). This indicates that submergence-induced hypoxia leads to ERF66 and ERF67 stabilization, similar to Arabidopsis ERFVIIs. Collectively, our protein stability assays reveal that ERF66 and ERF67 are substrates of the N-end rule pathway, whereas SUB1A-1 is not.
ERF66/ERF67 and SUB1A-1 Form a Signaling Cascade to Regulate Downstream Submergence Responses.
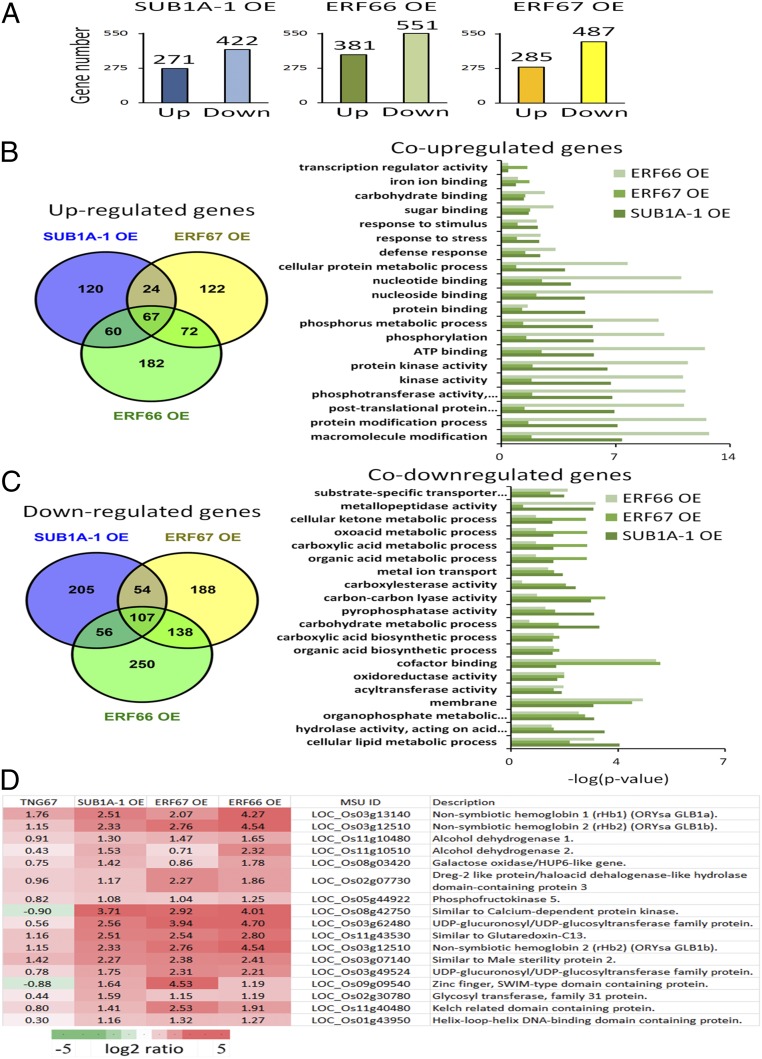
It is reported that SUB1A-1 could be activated by different abiotic stresses. We showed that ERF66 and ERF67 act genetically downstream of SUB1A-1 and are subjected to N-end rule regulation during hypoxia. Hence, we speculated that there are two sets of genes, one regulated by the SUB1A-1 and ERF66/ERF67 cascade and the other regulated solely by SUB1A-1. We carried out an RNA sequencing (RNA-seq) analysis of SUB1A-1 (line 1), ERF66 (line 2), and ERF67 (line 2) transgenic lines (as used in Fig. 3) to examine the effect of overexpressing these transcription factors on global gene expression (Dataset S1 shows full results). In these analyses, we first normalized expression levels after 24-h treatment of submergence with corresponding expression levels at 0 h treatment, and then eliminated any changes observed in the submergence-sensitive TNG67 background line. In doing so, we identified 271, 285, and 381 genes that were more than twofold up-regulated in the SUB1A-1, ERF67, and ERF66 individual-overexpression lines, respectively, but not in TNG67. Interestingly, ERF66 and ERF67 were approximately 3- and 30-fold up-regulated in the SUB1A-1 overexpression line, respectively. Furthermore, 422, 487, and 511 genes were more than twofold down-regulated in the SUB1A-1, ERF67, and ERF66 individual overexpression lines, respectively (Fig. 5A). Analysis of the gene lists revealed two distinct groupings of differentially expressed genes, one that is dependent on SUB1A-1 with ERF66 and/or ERF67 and the other that is regulated solely by SUB1A-1 (Fig. 5 B and C). For the first group, Venn diagram analyses show that 151 genes in total were up-regulated in SUB1A-1 and ERF67 (24 genes), SUB1A-1 and ERF66 (60 genes), and all three individual overexpression lines (67 genes), indicating that ERF66 and ERF67 may have different downstream targets (Fig. 5B). Meanwhile, we also found 217 genes in total that were down-regulated in SUB1A-1 and ERF67 (54 genes), SUB1A-1 and ERF66 (56 genes), and all three individual overexpression lines (107 genes; Fig. 5C).
Fig. 5.
Transcriptomic analyses of SUB1A-1, ERF66, and ERF67 overexpression lines under submergence. (A) Histogram showing numbers of up- and down-regulated genes (greater than twofold; P < 0.05) in SUB1A-1/ERF67/ERF66 overexpression lines that are not differentially regulated in TNG67. (B) Overlap among genes significantly up-regulated by overexpressing SUB1A-1, ERF66, and ERF66 and distribution of functional categories of up-regulated genes. (C) Overlap among genes significantly down-regulated by SUB1A-1, ERF66, and ERF67 and distribution of functional categories of down-regulated genes. Histograms in C and D indicate P values of the enriched functional categories. (D) Representative genes are up-regulated in SUB1A-1 and ERF66/67 overexpression lines. The first seven genes are orthologous of core hypoxia genes, which are up-regulated in SUB1A-1 and ERF66/67 overexpression lines.
A Gene Ontology analysis revealed that up-regulated genes were involved in diverse processes, including response to stress, defense response, phosphorylation, and protein kinase activity (Fig. 5B), whereas down-regulated genes included those associated with carbohydrate metabolic process and cellular lipid metabolic process (Fig. 5C). The up-regulated genes across all three transgenic lines included orthologs of core hypoxia-related genes, including nonsymbiotic hemoglobin 1/2 (LOC_Os03g13140/LOC_Os03g12510), alcohol dehydrogenase 1/2 (LOC_Os11g10480/LOC_Os11g10510), galactose oxidase/HUP6-like gene (LOC_Os08g03420), Dreg-2 like protein/haloacid dehalogenase-like hydrolase domain-containing protein 3 (LOC_Os02g07730), and phosphofructokinase 5 (LOC_Os05g44922; Fig. 5D and Dataset S1). This provides a transcriptional explanation for the enhanced submergence tolerance of these overexpression lines (Fig. 3), and further confirms the involvement of all three ERFVIIs in coordinating submergence responses.
The N Terminus of SUB1A-1 Has Random Coil Structure.
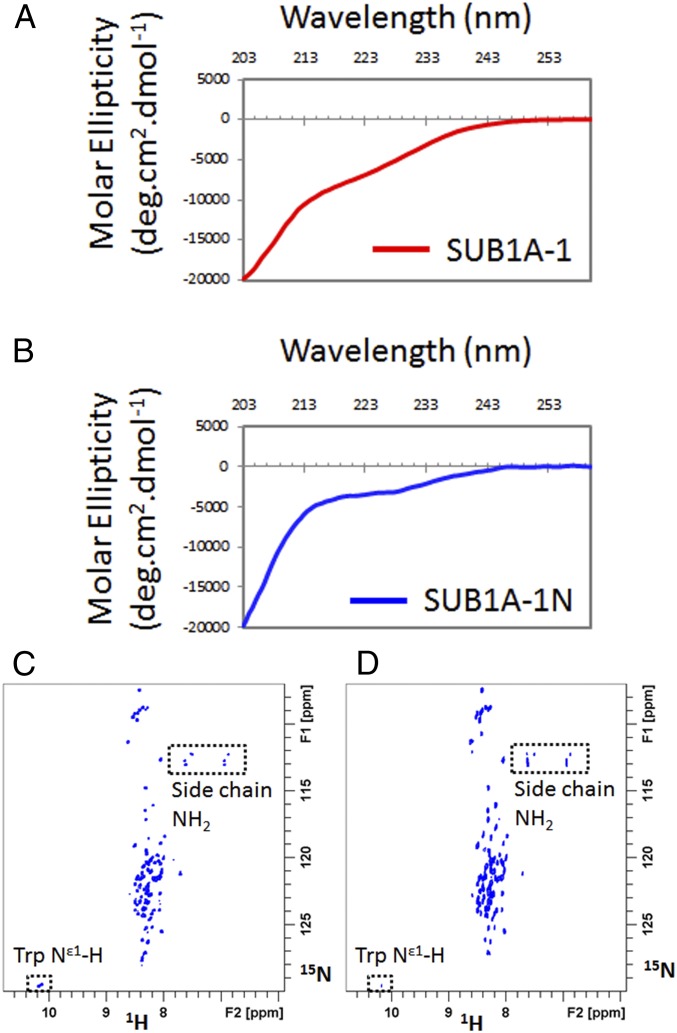
The key enzymes in the N-end rule pathway, including MetAP, ATE, and PRT6, are highly conserved in eukaryotes (22, 23). The active binding site of the human PRT6 functional homolog, UBR1, is a shallow and mostly hydrophobic pocket into which activated N-degrons can fit (34, 35). To understand how SUB1A-1 might evade N-end rule regulation despite having an N-terminal motif similar to the AtERFVIIs and OsERF66/67, we used CD and NMR spectroscopy to examine recombinant SUB1A-1 and SUB1A-1 N terminus (SUB1A-1N), which consists of the first 115 aa of SUB1A-1 (SI Appendix, Fig. S9) with an additional N-terminal serine (the residue of TEV protease cleavage site) and C-terminal His-tag for protein production and purification purposes. The secondary structure investigation by CD revealed that the full-length SUB1A-1 is mostly unstructured, and SUB1A-1N also resembles a random coil (Fig. 6 A and B). We further analyzed the structural properties of SUB1A-1N by NMR spectroscopy. As shown in Fig. 6C, the 2D 1H,15N–band-selective excitation short-transient (BEST)–heteronuclear single quantum coherence (HSQC) spectrum shows that the cross peaks of the backbone N-H groups of SUB1A-1N occur in a very narrow chemical shift range, indicative of a random coil structure when combining this result with its random coil CD curve. Furthermore, the solvent-exposed amide proton 2D 1H,15N-HSQC spectrum of SUB1A-1N shows that most of the amide protons have exchanged cross peaks with water (with an exchange rate greater than 3 Hz), indicating that backbone amides are solvent-exposed and are not protected by structure or hydrogen bonds (Fig. 6D). The combined CD and NMR analyses therefore indicate that SUB1A-1N is unstructured, suggesting that the N terminus of SUB1A-1 is very flexible and should be recognized by components of N-end rule. This raises the possibility that other regions of SUB1A-1 might be involved in preventing degradation by the N-end rule pathway or that other proteins bind to SUB1A-1 to shield the N-degron.
Fig. 6.
The CD and NMR spectra of recombinant SUB1A-1 constructs. CD spectra of full-length SUB1A-1 (A) and SUB1A-1 N terminus only (B) show that SUB1A-1 and its N terminus are mostly unstructured. The cross-peaks of 2D 1H,15N-BEST-HSQC spectrum of SUB1A-1 N terminus only occur in a very narrow chemical shift range (C), indicating a random coil structure when it combines with the CD result. The solvent-exposed amide proton 2D 1H,15N-HSQC spectrum of SUB1A-1 N terminus only (D) shows that most of the amide protons have exchange cross-peaks with water, indicating that amides are solvent-exposed.
The C Terminus of SUB1A-1 Prevents Its Degradation by the N-End Rule Pathway.
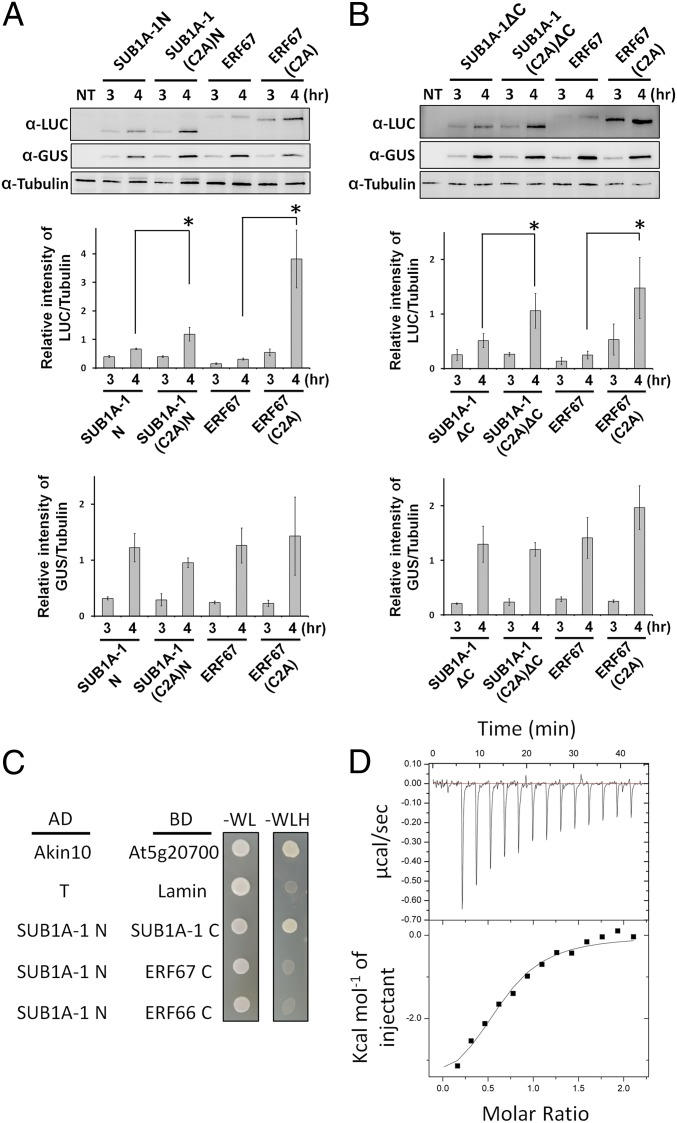
To test whether other regions of SUB1A-1 might interfere with its degradation by the N-end rule, we analyzed the protein stability of two C-terminally truncated variants of SUB1A-1 in TNG67 rice protoplast, including (i) SUB1A-1N (i.e., SUB1A-1 N terminus only) and (ii) SUB1A-1ΔC (SUB1A-1 lacking the C terminus; SI Appendix, Fig. S7B). The protein levels of SUB1A-1N and SUB1A-1ΔC were similar to WT ERF67 but significantly lower than ERF67(C2A), indicating that truncated SUB1A-1 is unstable after removing the C terminus (Fig. 7 A and B). To confirm that this instability is caused by degradation via the N-end rule pathway, we transiently expressed C2A variants (SI Appendix, Fig. S7B) of both truncation constructs in TNG67 rice protoplast cells. The protein quantities of SUB1A-1(C2A)N and SUB1A-1(C2A)ΔC were much higher than SUB1A-1N and SUB1A-1ΔC and similar to ERF67(C2A) (Fig. 7 A and B). This suggests that, in contrast to full-length SUB1A-1 (SI Appendix, Fig. S7C), C-terminally truncated variants of SUB1A-1 are degraded via N-end rule pathway. To understand how the SUB1A-1 C terminus interferes with SUB1A-1 degradation, we examined the capacity for these two regions of SUB1A-1 to interact with each other. Yeast two-hybrid analysis revealed an interaction between the SUB1A-1 N terminus and C terminus (Fig. 7C). This was specific for the SUB1A-1 C terminus, as ERF66 and ERF67 C termini did not interact with the SUB1A-1 N terminus. This interaction was also confirmed by isothermal titration calorimetry (iTC) experiments using recombinant SUB1A-1 N terminus and C terminus (Fig. 7D). Thus, we propose that the C-terminal region of SUB1A physically interacts with the SUB1A-1 N terminus, and that this shields the N-degron, preventing protein turnover.
Fig. 7.
C-terminally truncated SUB1A-1 can be degraded by the N-end rule pathway. Western blot analysis of protein stability of the truncated SUB1A-1 in TNG67 rice protoplasts. (A and B) Protein stability assays of SUB1A-1N and SUB1A-1ΔC, respectively. UbiP::SUB1A-1N/SUB1A-1ΔC/ERF67-Luc constructs were cotransfected into TNG67 rice protoplasts with a UbiP::GUS plasmid, which was used as a stable control. The transfected protoplasts were incubated in W5 solution for 3 or 4 h and then harvested for further Western blot analyses, and the relative levels of Luc and GUS were normalized to tubulin, respectively. NT, nontransfected. The data represent means ± SD from three independent replicates (*P < 0.05 indicates a significant difference by Student’s t test). The molecular weight of SUB1A-1N/SUB1A-1ΔC-Luc fusion proteins are ∼74/81 kDa, respectively. The molecular weight of the ERF67-Luc fusion protein is ∼85 kDa. (C) Yeast two-hybrid assay testing interactions between the SUB1A-1 N terminus and SUB1A-1, ERF66, or ERF67 C termini. AD, GAL4 activation domain; BD, GAL4 DNA binding domain. Positive interactions are represented by growth on the triple-dropout medium (-WLH), which tests for expression of the HIS3 reporter gene. Yeast growth on the double-dropout medium (-WL) is included as a cotransformation control. (D) iTC experiment for the binding between SUB1A-1 N and C termini. The upper curve shows corrected heat pulses resulting from titration of SUB1A-1 C terminus, and the lower graph shows the integrated heat pulse along with a fit.
Discussion
ERFVII transcription factors are involved in hypoxia-sensing and -regulating responses to flooding and/or hypoxic stress. For example, all five ERFVIIs in Arabidopsis function as important regulators of flooding and/or hypoxia tolerance, and ERFVIIs in barley, Rumex, and Rorippa regulate the response to waterlogging (12, 15, 17, 29, 36–40). Furthermore, Arabidopsis ERFVIIs have also been linked to other abiotic and biotic responses (12, 41–43). In rice, the ERFVII SUB1A-1 is the master regulator of the quiescence submergence-survival response, as well as other abiotic stresses (27, 28). However, in contrast to all other investigated ERFVIIs, SUB1A-1 was shown to resist the N-end rule pathway, suggesting that it is not directly involved in hypoxia sensing. In addition, SUB1A-1 does not confer flooding tolerance in Arabidopsis, suggesting some degree of difference between rice and Arabidopsis quiescence mechanism in response to submergence stresses. Here, we propose a regulatory cascade in the SUB1A-1–dependent submergence response that involves two other rice ERFVIIs, ERF66 and ERF67.
We dissected the transcriptional kinetics of 16 OsERFVIIs in two indica cultivars (submergence-tolerant FR13A and submergence-sensitive IR29) and found that many are transcriptionally up-regulated in response to submergence treatment (SI Appendix, Fig. S1). This includes ERF70, which was recently shown to contribute to improved recovery from submergence stress (44). Five of these OsERFVIIs (ERF59, ERF60, ERF61, ERF66, and ERF67) had higher transcript levels in FR13A than in IR29, and their expression patterns were similar to those of SUB1A-1 in FR13A and IR29 (SI Appendix, Fig. S1). By cross-examining the transcript levels of these five ERFVIIs in other submergence-tolerant cultivars, IR64(Sub1) and Swarna(Sub1), and other sensitive cultivars, IR64 and Swarna, we found that only ERF66 and ERF67 showed enhanced transcript abundance in tolerant cultivars than in sensitive cultivars (Fig. 1), indicating that they are downstream targets of SUB1A-1 during submergence.
By using trans-activation assays, we found that SUB1A-1 could transcriptionally activate ERF66 and ERF67 (Fig. 2 A and B). We also confirmed that SUB1A-1 can interact with ERF66 and ERF67 promoter by ChIP-qPCR (Fig. 2C). Multiple GCC boxes with different flanking sequences in the promoter regions of ERF66 and ERF67 are identified (SI Appendix, Fig. S4B), and our EMSA studies showed that recombinant SUB1A-1 selectively binds to several (but not all) of the identified GCC boxes (Fig. 2D and SI Appendix, Fig. S4C). Collectively, our data suggest that SUB1A-1 directly up-regulates ERF66 and ERF67 via interaction with GCC boxes in their promoters, and ERF66 and ERF67 are therefore downstream targets of SUB1A-1. Moreover, overexpression of ERF66 or ERF67 in the TNG67 submergence-sensitive cultivar led to enhanced submergence tolerance (Fig. 3).
By performing protein stability studies (Fig. 4 and SI Appendix, Figs. S7 and S8), we showed that ERF66 and ERF67, but not SUB1A-1 (19), are substrates for the N-end rule pathway, despite all three proteins having canonical N-degron sequences in their N termini. The NMR analyses showed that the N terminus of SUB1A-1 is a random coil structure (Fig. 6), indicating that the N terminus of SUB1A-1 is very flexible and should be easily recognized by the N-end rule-related enzymes. This raised the question of how SUB1A-1 can escape N-end rule regulation.
Our assays (Fig. 7 A and B) showed that C-terminally truncated SUB1A-1 could be degraded via N-end rule pathway, suggesting that the C terminus of SUB1A-1 is involved in inhibiting its degradation. Yeast two-hybrid and iTC experiments showed the physical interaction between N and C termini of SUB1A-1 (Fig. 7 C and D). Hence, it is likely that C terminus of SUB1A-1 helps mask the N-terminal region involved in the N-end rule pathway. An analogous scenario has been reported for the α-synuclein protein, whereby long-range interdomain interactions lead to stabilization by adopting an ensemble of conformations to mask its amyloidogenic domain (45, 46). Taken together, these results suggest that features in the N terminus and C terminus of SUB1A-1 contribute to its escape from N-end rule degradation, likely through domain–domain interactions that prevent adequate exposure of the N terminus or block the site of ubiquitination. However, the detailed molecular mechanism remains unclear and requires further investigation.
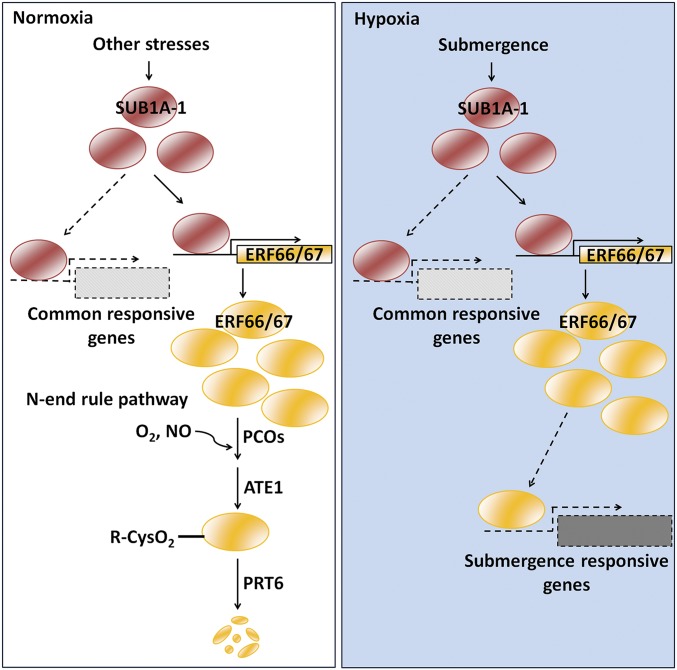
SUB1A-1 is a major factor that confers submergence tolerance in rice. It is up-regulated not only under submergence, but also during drought, prolonged darkness, oxidative stress, and ethylene stress, and plays a key role in a range of abiotic stress responses in addition to submergence (8, 27, 28, 47, 48). The decoupling of SUB1A-1 from N-end rule regulation may have allowed SUB1A-1 to adopt a wider range of functions as a master transcriptional regulator, functioning as a hub to orchestrate the signaling networks in response to various stresses under normoxic or hypoxic conditions. However, this raised the question of how rice discriminates the submergence stress (hypoxia) from other SUB1A-1–regulated stresses that occur when oxygen is readily available. In this study, we identified ERF66 and ERF67 as direct downstream targets of SUB1A-1 and substrates of the N-end rule pathway, which may be critical for coordinating hypoxia responses. In addition, RNA-seq analyses showed that two distinct groups gene were induced by SUB1A-1; one group is dependent on ERF66 and ERF67 and the other group is independent of ERF66 and ERF67 (Fig. 5 and Dataset S1). SUB1A-1 is a transcription factor involving several important processes under submergence stress (49). Our data suggested that ERF66 and ERF67 are the downstream genes of SUB1A-1, and these two genes are involved in several important processes during submergence and confer submergence tolerance to rice as well as SUB1A-1. This appears similar to the situation in Arabidopsis, in which the ERFVIIs HRE1 and HRE2 are downstream of RAP2.2, 2.3, and 2.12 (15). We propose that this SUB1A-1–to–ERF66/ERF67 regulatory cascade is the link that allows rice to distinguish between submergence and other abiotic stresses (Fig. 8). In this model, SUB1A-1 is induced under different abiotic stresses, which in turn activates ERF66/ERF67 genes and a set of common stress response genes. Under normoxic abiotic stress conditions, ERF66 and ERF67 are degraded via the N-end rule pathway. Only under low oxygen conditions would ERF66 and ERF67 be stabilized, accumulating to trigger hypoxic responses, and allowing FR13A and flooding-tolerant cultivars to survive as long as 2 wk under complete submergence. The constitutive stability of SUB1A-1 means that, when oxygen levels have returned to normal after desubmergence, ERF66/67 would be quickly degraded to switch off the specific hypoxia transcriptional response, but SUB1A-1 would remain stable to coordinate the expression of other genes that are needed for desubmergence/drought/ROS survival.
Fig. 8.
Model of the regulatory cascade of SUB1A-1, ERF66, and ERF67 that involves transcriptional and N-end rule pathways in response to submergence stress in submergence-tolerant rice cultivars.
Materials and Methods
Further details of experimental procedures are provided in SI Appendix, SI Materials and Methods.
Plant Materials.
Rice (O. sativa) cultivars FR13A, IR29, IR64, Swarna, and TNG67 were used in this study. Two near-isogenic lines, IR64(Sub1) and Swarna(Sub1), were provided by the National Plant Genetic Resources Center of the Taiwan Agricultural Research Institute (Taichung City, Taiwan). SUB1A-1/ERF66/ERF67 overexpression transgenic rice lines (SUB1A-1 OE/ERF66 OE/ERF67 OE) were generated by transforming the UbiP::GST-SUB1A-1/ERF66/ERF67 in pCAMBIA1301 vector into TNG67 rice. ERF66 and ERF67 overexpression transgenic Arabidopsis lines were generated by transforming the 35S::ERF66/ERF67-GFP in the pK7FWG2 vector into Arabidopsis (A. thaliana) ecotype Columbia-0. Transformation into Agrobacterium tumefaciens and Arabidopsis was performed according to established protocols (50). Rice transformation was done by the Transgenic Plant Core Lab of Academia Sinica.
Growth Conditions and Submergence Treatment.
Rice seeds were sterilized with 1.2% (vol/vol) sodium hypochlorite containing 0.1% (vol/vol) Tween 20 for 30 min and washed at least five times with sterilized water. The sterilized seeds were placed on moist filter paper in Petri dishes at 37 °C in the dark for 4 d. After incubation, uniformly germinated seeds were transplanted onto an iron grid in a beaker with quarter-strength Kimura B solution, pH 5.6–5.8 (51), and the solution was renewed every 2 d. The hydroponically cultivated seedlings were grown in a growth chamber at 28 °C with a 16-h-light (120–125 μmolˑm−2ˑs−1)/8-h-dark cycle until they were 14 d old. For submergence treatment of 14-d-old rice seedlings, beakers with plants were placed into a water tank (40 cm wide × 40 cm long × 70 cm tall) filled 55 cm high with tap water for the indicated times at 28 °C in the dark. For phenotypic assays, data were collected from each genotype in two independent experiments. Fourteen-day-old seedlings were subjected to submergence treatment as previously described for 7 d in the dark. After submergence treatment, the rice seedlings were put back into the growth chamber at 28 °C with a 16-h-light (120–125 μmolˑm−2ˑs−1)/8-h-dark cycle for a further 14 d of recovery, followed by evaluation of whole plant viability. Plants were scored as viable when one or more new leaves appeared during the recovery period.
Protoplast Preparation and Transformation.
Rice protoplast preparation and transformation were conducted as described (52 and 53) with minor modifications. For protoplast preparation, the stems and sheaths of the 14-d-old TNG67 rice seedlings were cut into 0.5-mm strips and incubated in an enzyme solution [2% cellulase RS (Yakult), 1% macerozyme R10 (Yakult), 0.1% Mes, pH 5.6, 0.6 M mannitol, 0.1% CaCl2, and 1% BSA] and vacuum-infiltrated (15–20 cm Hg) for 15 min. After vacuum infiltration, the strips in the enzyme solution were gently shaken under light for approximately 3.5 h until the protoplasts were released into the solution. After digestion, the solutions containing protoplasts were filtered through 40-μm nylon meshes, followed by centrifugation at 200 × g for 3 min with a swinging bucket to pellet the protoplasts in a round-bottomed tube. The supernatants were removed, the protoplast pellets were resuspended in W5 solution [154 mM NaCl, 125 mM CaCl2, 5 mM KCl, 5 mM glucose, and 2 mM 4-morpholineethanesulfonic acid (MES), pH 5.7], and this step was repeated once before protoplasts were incubated and resuspended on ice for at least 30 min. Then, the W5 solution was removed, and protoplasts were resuspended to a final concentration of 2–5 × 105 cells per milliliter in MMG solution (0.6 M mannitol, 15 mM MgCl2, and 4 mM MES, pH 5.7). For protoplast transformation, a total of approximately 4 × 105 protoplasts in 0.2 mL of MMG solution were mixed with 20 μg of plasmid DNA on ice for 10 min. Then, an equal volume (approximately 220 μL) of PEG–calcium solution [40% wt/vol PEG 4000 (95904; Sigma-Aldrich), 0.6 M mannitol, and 0.1 M CaCl2] was added, and the mixture was incubated at room temperature (RT) for 20 min. After incubation, 3 mL of W5 solution was added slowly and gently mixed, and the protoplasts were pelleted by centrifugation at 200 × g for 1 min with a swinging bucket. After washing twice with W5 solution, the pellets were resuspended gently in 1.5 mL of W5 solution and incubated in six-well plates coated with 1% BSA at RT for the indicated times in dark.
ChIP-qPCR Assay.
To detect direct target genes of SUB1A-1, ChIP-qPCR assays were performed by using a rice protoplast system. Cross-linking was conducted as described (54) with minor modification. Briefly, UbiP::SUB1A-1-Luc constructs and Ubi::Luc constructs (as a control) were transfected into TNG67 rice protoplasts, respectively. After 4 h incubation at RT, the transfected protoplasts (1.6 × 106 cells per transfection) were collected by centrifugation at 200 × g for 2 min at RT, followed by removal of the supernatants. The collected protoplasts were subjected to cross-linking with 1% formaldehyde in 1.5 mL W5 solution and gently mixed on a rotor (12 rpm) for 10 min at RT. To quench the cross-linking reaction, 80 μL of 2M glycine was added and gently mixed on a rotor (Intelli Mixer ERM-2L; ELMI) at 12 rpm for 5 min at RT, followed by centrifugation at 1,500 × g for 5 min at 4 °C to remove the supernatant, and the protoplasts were rinsed with 1 mL of ice-cold 1× PBS buffer (pH 7.4). Chromatin extraction, MNase digestion, sonication, immunoprecipitation, reverse cross-linking, recovery of DNA, and qPCR were conducted by using a Pierce Magnetic ChIP kit (cat. no. 26157; Thermo Scientific). These procedures followed the protocol provided by the manufacturer. The DNA–protein complex was immune-precipitated with anti-Luciferase antibody (sc-74548; Santa Cruz) at a concentration of 5 μg for each immunoprecipitation. The bound DNA fragments were then reversely released and amplified by specific qPCR reaction. The primers used in ChIP-qPCR assay are listed in SI Appendix, Table S1.
In Vitro Analyses of Protein Stability.
In vitro analyses of protein stability was conducted as described previously (19). A modified version of pTNT (Invitrogen) expression vector, pTNT4×MYC, which possesses T7 promoter and SP6 promoter, 5 β-globin leader, ccdB fragment, 4×MYC fragment, and T7 terminator sequentially, was generated to perform the in vitro analysis. First, pTNT was double-digested with XhoI and XbaI and gel-eluted to purify the sticky-ended pTNT vector. The pGWB516 plasmid was used as a template to amplify ccdB fragments carrying XhoI and EcoRV site at the 5′ and 3′ end and 4×MYC fragments carrying EcoRV and XbaI site at the 5′ and 3′ end by PCR, respectively. The primers used here are listed in SI Appendix, Table S2. The amplified ccdB fragments were then double-digested with XhoI and EcoRV, and the amplified 4×MYC fragments were double-digested with EcoRV and XbaI. After gel elution, the ccdB fragment and 4×MYC fragment were coligated into the sticky-ended pTNT vectors to generate the pTNT4×MYC. The details on sequence of pTNT4×MYC are shown in SI Appendix, Fig. S10. The CDS of ERF66 and ERF67 were cloned from cDNA derived from submerged FR13A cDNA, and the ERF66 and ERF67 DNA fragments were subcloned into pTNT4×MYC by Gateway system (Invitrogen) to produce C-terminal MYC-tagged fusions driven by T7 promoter. The CDS of SUB1A-2 was cloned from cDNA derived from submerged IR29 cDNA and ligated into the modified pTNT3×HA (19) to produce C-terminal HA-tagged fusions driven by T7 promoter. The pTNT3×HA-SUB1A-1 was from Gibbs et al. (19). N-terminal mutations were incorporated by changing the forward primer sequences accordingly (SI Appendix, Table S2). In vitro assays of protein stability were carried out by using rabbit reticulocyte lysate system (L4960; Promega) with the addition of 100 μM CHX to block mRNA translation. Reactions were first incubated for 30 min at 30 °C to allow protein translation. Following this 30-min period, CHX is added to prevent further translation and a sample of the reactions is taken immediately (i.e., at time 0), and then the following samples were taken at indicated time points before mixing with protein loading dye to terminate protein synthesis. Equal amounts of each reaction were subjected to anti-HA/MYC immunoblot analysis. All blots were checked for equal loading by Ponceau staining.
Western Blot Analysis and Antibodies.
Protein extraction from the transfected rice protoplasts (4 × 105 cells per transfection) was conducted as described (55). Protein extraction from the transgenic Arabidopsis seedling was conducted as described (56). Proteins resolved by SDS/PAGE were transferred to PVDF by using a MiniTrans-Blot electrophoretic transfer cell (Bio-Rad). Membranes were probed with primary antibody at the following titers: anti-HA (H3663; Sigma-Aldrich), 1:1,000; anti-MYC (WH0004609M2; Sigma-Aldrich), 1:1,000; anti-tubulin (T5168; Sigma-Aldrich), 1:5,000; anti-GUS (G5545; Sigma-Aldrich), 1:1,000; and anti-Luciferase (sc-74548; Santa Cruz), 1:200. HRP-conjugated anti-mouse (NEF822001EA; PerkinElmer)/rabbit (DC03L; Calbiochem) secondary antibody was used at a titer of 1:3,000. Immunoblots were visualized with enhanced chemiluminescence reagent (SuperSignal West Pico; Thermo Scientific). The relative image intensities were quantified by using ImageJ (https://imagej.nih.gov/ij/).
Protein Expression and Purification.
The pET32a-SUB1A-1 and pET32a-SUB1A-1N were transformed into Escherichia coli Rosetta (DE3). Recombinant protein expression was induced at OD 0.6 by adding 1 mM IPTG at 25 °C for 6 h and harvested by centrifugation at 4,700 × g for 30 min. Cells were resuspended in lysis buffer A (50 mM Hepes, pH 7.5, 400 mM NaCl, and 20 mM imidazole) supplemented with 10 μg/mL of DNase I, 1 mg/mL of lysozyme, and 1 mM PMSF and lysed by sonication. The lysate was centrifuged at 20,000 × g for 30 min, and the supernatant was loaded onto a column containing NiNTA resin preequilibrated with lysis buffer. The column was washed with 20 column volumes (CVs) of lysis buffer followed by 5 CVs of wash buffer (50 mM Hepes, pH 7.5, 300 mM NaCl, and 50 mM imidazole). Proteins were eluted with elution buffer (50 mM Hepes, pH 7.5, 100 mM NaCl, 500 mM imidazole). After removing thioredoxin tag by overnight TEV protease treatment at 4 °C, the solution was loaded onto a HiPrep Heparin FF 16/10 column preequilibrated with buffer A (50 mM Hepes, pH 7.5, 10 mM β-mercaptoethanol, and 5% glycerol) with 100 mM NaCl. After column washing, the protein was eluted with a 0–100% gradient of buffer A with 1 M NaCl in 20 CVs. The protein containing fractions of the major peak were concentrated and polished by using an ENrich SEC650 column with buffer A with 300 mM NaCl.
For 15N-labeled SUB1A-1 N terminus, E. coli Rosetta (DE3) was cultured in Luria broth until OD reached 1.0 and centrifuged at 4,000 × g for 20 min. Cell pellets were then washed and resuspended in M9 buffer three times. Resuspended cells were recovered in M9 medium with [15N]NH4Cl as the sole nitrogen source for 1 h at 37 °C before overnight induction at 16 °C by adding 0.5 mM IPTG.
CD Spectrometry.
The far-UV CD spectra were recorded over a range of 204–260 nm at 25 °C by using a Jasco J-815 spectrometer (Jasco). A total of 5.5 μM of SUB1A-1 and 12 μM of SUB1A-1N (both in 5 mM Hepes, pH 7.5, and 100 mM NaCl) were transferred to a 1-mm quartz cuvette before data collection. All spectra were buffer-subtracted and smoothed by using Spectra Analysis (Jasco). The results are expressed as the mean residual molar ellipticity.
NMR Spectroscopy.
The NMR sample in a 4-mm OD Shigemi tube contained 200 μL, pH 7.0, aqueous buffer solution (90% H2O/10% D2O) with 0.14 mM 15N-labeled SUB1A-1N protein, 150 mM NaCl, 25 mM potassium phosphate, 1 mM NaN3, and 0.1 mM 4,4-dimethyl-4-silapentane-1-sulfonic acid as an internal chemical shift standard. All NMR data were collected at 298 K on a Bruker 800 MHz NMR spectrometer (AV800) equipped with a TXI cryogenic probe. Two-dimensional 1H,15N-HSQC spectra were collected with a BEST scheme (57). Acquisition parameters for BEST 2D 1H,15N-HSQC were as follows: the center of the N-H proton selective pulses at 8.5 ppm, 0.2-s interscan delay, 512 scans per increment, 256 increments in the 15N dimension accumulated. Solvent-exposed 2D 1H,15N-HSQC data were collected by using the Phase-Modulated CLEAN chemical EXchange scheme (58) with a 100-ms exchange mixing, 360 scans per increment, and 128 increments in the 15N dimension. NMR data were processed by using Topspin software (Bruker).
iTC Binding Assays.
The iTC experiment was performed on an ITC200 calorimeter (MicroCal) at 25 °C. The measurement buffer consisted of 50 mM Hepes, 100 mM NaCl, and 1 mM DTT, pH 8.0. The injection syringe (40 μL) was filled with 1 mM SUB1A-1 N terminus, and the sample cell was loaded with 200 μL of 100 μM SUB1A-1 C terminus. First injection (0.3 μL) of SUB1A-1 N domain was followed by 13 injections of 3 μL at a stirring speed of 1,000 rpm. The titration value of the first injection was not used in data analysis. The best fits to the binding isotherms were obtained by subtracting saturated integral of signal from the last point as reference. However, the interaction is relatively weak, so thermodynamic parameters (N, ΔH, KA) could not be accurately estimated. All data were plotted and analyzed by Microcal Origin software.
Details of RNA extraction and qRT-PCR, RNA-seq and data analysis, plasmid construction, trans-activation assay, EMSA, and yeast two-hybrid assay are described in SI Appendix, SI Materials and Methods. All of the primers for qRT-PCR and cDNA cloning and the probes for EMSA are listed in SI Appendix, Tables S1–S3.
Supplementary Material
Acknowledgments
We thank the Plant Tech Core Facility, Genomic Technology Core - Microarray and Sequencing Division of the Institute of Plant and Microbial Biology, the Transgenic Plant Core Lab, and the Academia Sinica Biotechnology Center in Southern Taiwan (AS-BCST) greenhouse core facility of Academia Sinica for technical support. We acknowledge the use of the instruments in the Biophysics Core Facility, Scientific Instrument Center at Academia Sinica. We thank Academia Sinica High-Field NMR Center (HFNMRC) for technical support; HFNMRC is funded by Academia Sinica Core Facility and Innovative Instrument Project (AS-CFII-108-112). This work was supported by Academia Sinica and Ministry of Science and Technology Grant MOST 107-2311-B-001-002 (to M.-C.H.), Taiwan Protein Project Grant AS-KPQ-105-TPP (to M.-C.H.), Biotechnology and Biological Scientific Research Council Grant BB/M020568/1 (to D.J.G.), and European Research Council Starting Grant 715441-GasPlaNt (to D.J.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the National Center for Biotechnology Information BioProject database, https://www.ncbi.nlm.nih.gov/bioproject (accession no. PRJNA512592).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1818507116/-/DCSupplemental.
References
- 1.Sasidharan R, Voesenek LACJ. Ethylene-mediated acclimations to flooding stress. Plant Physiol. 2015;169:3–12. doi: 10.1104/pp.15.00387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailey-Serres J, et al. Making sense of low oxygen sensing. Trends Plant Sci. 2012;17:129–138. doi: 10.1016/j.tplants.2011.12.004. [DOI] [PubMed] [Google Scholar]
- 3.Gibbs J, Greenway H. Mechanism of anoxia tolerance in plant. I. Growth, survival and anaerobic catabolism. Funct Plant Biol. 2003;30:1–47. doi: 10.1071/PP98095. [DOI] [PubMed] [Google Scholar]
- 4.Hattori Y, et al. The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature. 2009;460:1026–1030. doi: 10.1038/nature08258. [DOI] [PubMed] [Google Scholar]
- 5.Xu K, et al. Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature. 2006;442:705–708. doi: 10.1038/nature04920. [DOI] [PubMed] [Google Scholar]
- 6.Kuroha T, et al. Ethylene-gibberellin signaling underlies adaptation of rice to periodic flooding. Science. 2018;361:181–186. doi: 10.1126/science.aat1577. [DOI] [PubMed] [Google Scholar]
- 7.Singh P, Sinha AK. A positive feedback loop governed by SUB1A1 interaction with mitogen-activated protein kinase3 imparts submergence tolerance in rice. Plant Cell. 2016;28:1127–1143. doi: 10.1105/tpc.15.01001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fukao T, Xu K, Ronald PC, Bailey-Serres J. A variable cluster of ethylene response factor-like genes regulates metabolic and developmental acclimation responses to submergence in rice. Plant Cell. 2006;18:2021–2034. doi: 10.1105/tpc.106.043000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh S, Mackill DJ, Ismail AM. Responses of SUB1 rice introgression lines to submergence in the field: Yield and grain quality. Field Crops Res. 2009;113:12–23. [Google Scholar]
- 10.Nakano T, Suzuki K, Fujimura T, Shinshi H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006;140:411–432. doi: 10.1104/pp.105.073783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bui LT, Giuntoli B, Kosmacz M, Parlanti S, Licausi F. Constitutively expressed ERF-VII transcription factors redundantly activate the core anaerobic response in Arabidopsis thaliana. Plant Sci. 2015;236:37–43. doi: 10.1016/j.plantsci.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 12.Gibbs DJ, et al. Group VII ethylene response factors coordinate oxygen and nitric oxide signal transduction and stress responses in plants. Plant Physiol. 2015;169:23–31. doi: 10.1104/pp.15.00338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hinz M, et al. Arabidopsis RAP2.2: An ethylene response transcription factor that is important for hypoxia survival. Plant Physiol. 2010;153:757–772. doi: 10.1104/pp.110.155077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hess N, Klode M, Anders M, Sauter M. The hypoxia responsive transcription factor genes ERF71/HRE2 and ERF73/HRE1 of Arabidopsis are differentially regulated by ethylene. Physiol Plant. 2011;143:41–49. doi: 10.1111/j.1399-3054.2011.01486.x. [DOI] [PubMed] [Google Scholar]
- 15.Licausi F, et al. HRE1 and HRE2, two hypoxia-inducible ethylene response factors, affect anaerobic responses in Arabidopsis thaliana. Plant J. 2010;62:302–315. doi: 10.1111/j.1365-313X.2010.04149.x. [DOI] [PubMed] [Google Scholar]
- 16.Park HY, et al. AtERF71/HRE2 transcription factor mediates osmotic stress response as well as hypoxia response in Arabidopsis. Biochem Biophys Res Commun. 2011;414:135–141. doi: 10.1016/j.bbrc.2011.09.039. [DOI] [PubMed] [Google Scholar]
- 17.Yang CY, Hsu FC, Li JP, Wang NN, Shih MC. The AP2/ERF transcription factor AtERF73/HRE1 modulates ethylene responses during hypoxia in Arabidopsis. Plant Physiol. 2011;156:202–212. doi: 10.1104/pp.111.172486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sasidharan R, Mustroph A. Plant oxygen sensing is mediated by the N-end rule pathway: A milestone in plant anaerobiosis. Plant Cell. 2011;23:4173–4183. doi: 10.1105/tpc.111.093880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gibbs DJ, et al. Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nature. 2011;479:415–418. doi: 10.1038/nature10534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Licausi F, et al. Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nature. 2011;479:419–422. doi: 10.1038/nature10536. [DOI] [PubMed] [Google Scholar]
- 21.Gibbs DJ, et al. Nitric oxide sensing in plants is mediated by proteolytic control of group VII ERF transcription factors. Mol Cell. 2014;53:369–379. doi: 10.1016/j.molcel.2013.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graciet E, Mesiti F, Wellmer F. Structure and evolutionary conservation of the plant N-end rule pathway. Plant J. 2010;61:741–751. doi: 10.1111/j.1365-313X.2009.04099.x. [DOI] [PubMed] [Google Scholar]
- 23.Gibbs DJ, Bacardit J, Bachmair A, Holdsworth MJ. The eukaryotic N-end rule pathway: Conserved mechanisms and diverse functions. Trends Cell Biol. 2014;24:603–611. doi: 10.1016/j.tcb.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 24.Weits DA, et al. Plant cysteine oxidases control the oxygen-dependent branch of the N-end-rule pathway. Nat Commun. 2014;5:3425. doi: 10.1038/ncomms4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.White MD, et al. Plant cysteine oxidases are dioxygenases that directly enable arginyl transferase-catalysed arginylation of N-end rule targets. Nat Commun. 2017;8:14690. doi: 10.1038/ncomms14690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peña-Castro JM, et al. Expression of rice SUB1A and SUB1C transcription factors in Arabidopsis uncovers flowering inhibition as a submergence tolerance mechanism. Plant J. 2011;67:434–446. doi: 10.1111/j.1365-313X.2011.04605.x. [DOI] [PubMed] [Google Scholar]
- 27.Fukao T, Yeung E, Bailey-Serres J. The submergence tolerance regulator SUB1A mediates crosstalk between submergence and drought tolerance in rice. Plant Cell. 2011;23:412–427. doi: 10.1105/tpc.110.080325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fukao T, Yeung E, Bailey-Serres J. The submergence tolerance gene SUB1A delays leaf senescence under prolonged darkness through hormonal regulation in rice. Plant Physiol. 2012;160:1795–1807. doi: 10.1104/pp.112.207738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jung KH, et al. The submergence tolerance regulator Sub1A mediates stress-responsive expression of AP2/ERF transcription factors. Plant Physiol. 2010;152:1674–1692. doi: 10.1104/pp.109.152157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohme-Takagi M, Shinshi H. Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell. 1995;7:173–182. doi: 10.1105/tpc.7.2.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sessa G, Meller Y, Fluhr R. A GCC element and a G-box motif participate in ethylene-induced expression of the PRB-1b gene. Plant Mol Biol. 1995;28:145–153. doi: 10.1007/BF00042046. [DOI] [PubMed] [Google Scholar]
- 32.Allen MD, Yamasaki K, Ohme-Takagi M, Tateno M, Suzuki M. A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA. EMBO J. 1998;17:5484–5496. doi: 10.1093/emboj/17.18.5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gasch P, et al. Redundant ERF-VII transcription factors bind to an evolutionarily conserved cis-motif to regulate hypoxia-responsive gene expression in Arabidopsis. Plant Cell. 2016;28:160–180. doi: 10.1105/tpc.15.00866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choi WS, et al. Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases. Nat Struct Mol Biol. 2010;17:1175–1181. doi: 10.1038/nsmb.1907. [DOI] [PubMed] [Google Scholar]
- 35.Matta-Camacho E, Kozlov G, Li FF, Gehring K. Structural basis of substrate recognition and specificity in the N-end rule pathway. Nat Struct Mol Biol. 2010;17:1182–1187. doi: 10.1038/nsmb.1894. [DOI] [PubMed] [Google Scholar]
- 36.van Dongen JT, Licausi F. Oxygen sensing and signaling. Annu Rev Plant Biol. 2015;66:345–367. doi: 10.1146/annurev-arplant-043014-114813. [DOI] [PubMed] [Google Scholar]
- 37.Mendiondo GM, et al. Enhanced waterlogging tolerance in barley by manipulation of expression of the N-end rule pathway E3 ligase PROTEOLYSIS6. Plant Biotechnol J. 2016;14:40–50. doi: 10.1111/pbi.12334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lasanthi-Kudahettige R, et al. Transcript profiling of the anoxic rice coleoptile. Plant Physiol. 2007;144:218–231. doi: 10.1104/pp.106.093997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tamang BG, Magliozzi JO, Maroof MAS, Fukao T. Physiological and transcriptomic characterization of submergence and reoxygenation responses in soybean seedlings. Plant Cell Environ. 2014;37:2350–2365. doi: 10.1111/pce.12277. [DOI] [PubMed] [Google Scholar]
- 40.van Veen H, et al. Group VII ethylene response factor diversification and regulation in four species from flood-prone environments. Plant Cell Environ. 2014;37:2421–2432. doi: 10.1111/pce.12302. [DOI] [PubMed] [Google Scholar]
- 41.de Marchi R, et al. The N-end rule pathway regulates pathogen responses in plants. Sci Rep. 2016;6:26020. doi: 10.1038/srep26020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vicente J, et al. The Cys-Arg/N-end rule pathway is a general sensor of abiotic stress in flowering plants. Curr Biol. 2017;27:3183–3190.e4. doi: 10.1016/j.cub.2017.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vicente J, et al. Distinct branches of the N-end rule pathway modulate the plant immune response. New Phytol. 2019;221:988–1000. doi: 10.1111/nph.15387. [DOI] [PubMed] [Google Scholar]
- 44.Jisha V, et al. Overexpression of an AP2/ERF type transcription factor OsEREBP1 confers biotic and abiotic stress tolerance in rice. PLoS One. 2015;10:e0127831. doi: 10.1371/journal.pone.0127831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bertoncini CW, Fernandez CO, Griesinger C, Jovin TM, Zweckstetter M. Familial mutants of α-synuclein with increased neurotoxicity have a destabilized conformation. J Biol Chem. 2005;280:30649–30652. doi: 10.1074/jbc.C500288200. [DOI] [PubMed] [Google Scholar]
- 46.Ehrnhoefer DE, et al. EGCG redirects amyloidogenic polypeptides into unstructured, off-pathway oligomers. Nat Struct Mol Biol. 2008;15:558–566. doi: 10.1038/nsmb.1437. [DOI] [PubMed] [Google Scholar]
- 47.Alpuerto JB, Hussain RMF, Fukao T. The key regulator of submergence tolerance, SUB1A, promotes photosynthetic and metabolic recovery from submergence damage in rice leaves. Plant Cell Environ. 2016;39:672–684. doi: 10.1111/pce.12661. [DOI] [PubMed] [Google Scholar]
- 48.Tamang BG, Fukao T. Plant adaptation to multiple stresses during submergence and following desubmergence. Int J Mol Sci. 2015;16:30164–30180. doi: 10.3390/ijms161226226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Locke AM, Barding GA, Jr, Sathnur S, Larive CK, Bailey-Serres J. Rice SUB1A constrains remodelling of the transcriptome and metabolome during submergence to facilitate post-submergence recovery. Plant Cell Environ. 2018;41:721–736. doi: 10.1111/pce.13094. [DOI] [PubMed] [Google Scholar]
- 50.Hsu FC, et al. Submergence confers immunity mediated by the WRKY22 transcription factor in Arabidopsis. Plant Cell. 2013;25:2699–2713. doi: 10.1105/tpc.113.114447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoshida S, Forno DA, Cook JH, Gomez KA. 1976. Routine procedures for growing rice plants in culture solution in Laboratory Manual for Physiological Studies of Rice (International Rice Research Institute, Manila, Philippines). pp.61-66.
- 52.Wu FH, et al. Tape–Arabidopsis Sandwich–a simpler Arabidopsis protoplast isolation method. Plant Methods. 2009;5:16. doi: 10.1186/1746-4811-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Y, et al. A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods. 2011;7:30. doi: 10.1186/1746-4811-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee JH, Jin S, Kim SY, Kim W, Ahn JH. A fast, efficient chromatin immunoprecipitation method for studying protein-DNA binding in Arabidopsis mesophyll protoplasts. Plant Methods. 2017;13:42. doi: 10.1186/s13007-017-0192-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lu CA, et al. The SnRK1A protein kinase plays a key role in sugar signaling during germination and seedling growth of rice. Plant Cell. 2007;19:2484–2499. doi: 10.1105/tpc.105.037887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cho HY, Wen TN, Wang YT, Shih MC. Quantitative phosphoproteomics of protein kinase SnRK1 regulated protein phosphorylation in Arabidopsis under submergence. J Exp Bot. 2016;67:2745–2760. doi: 10.1093/jxb/erw107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lescop E, Schanda P, Brutscher B. A set of BEST triple-resonance experiments for time-optimized protein resonance assignment. J Magn Reson. 2007;187:163–169. doi: 10.1016/j.jmr.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 58.Hwang TL, van Zijl PCM, Mori S. Accurate Quantitation of Water-amide Proton Exchange Rates Using the Phase-Modulated CLEAN Chemical Exchange (CLEANEX-PM) Approach with a Fast-HSQC (FHSQC) Detection Scheme. J Biomol NMR. 1998;11:221–226. doi: 10.1023/a:1008276004875. [DOI] [PubMed] [Google Scholar]
- 59.Welsch R, Maass D, Voegel T, DellaPenna D, Beyer P. Transcription factor RAP2.2 and its interacting partner SINAT2: stable elements in the carotenogenesis of Arabidopsis leaves. Plant Physiol. 2007;145:1073–1085. doi: 10.1104/pp.107.104828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Prajapati GK, Kashyap N, Kumar A, Pandey DM. Identification of GCC box in the promoter region of ubiquinol cytochrome C chaperone gene using molecular beacon probe and its in silico protein-DNA interaction study in rice (Oryza sativa L.) Int J Comput Bioinfo In. Silico Model. 2013;2:213–222. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.