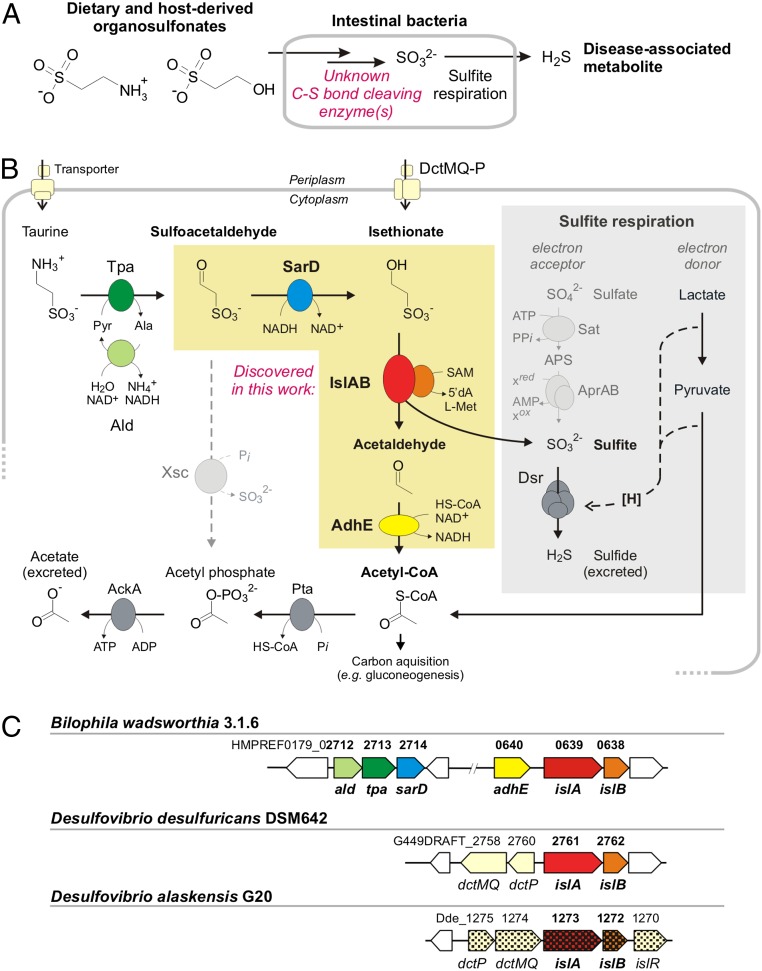
Fig. 1.
Metabolism of taurine and isethionate by the human gut bacterium B. wadsworthia and by Desulfovibrio spp. (A) B. wadsworthia and other intestinal bacteria degrade dietary and host-derived organosulfonates to access sulfite as an electron acceptor for their anaerobic respiration. The desulfonation reaction in B. wadsworthia has not yet been identified. (B) Summary of the pathways investigated in this study (no single organism represented). B. wadsworthia uses taurine and isethionate as electron acceptors, and the two Desulfovibrio spp. strains can use isethionate, but not taurine. All three strains lack the Xsc enzyme known in many aerobic bacteria (dashed line on the left). Instead, the GRE IslA with its GRE-activase component (IslB) are found in both B. wadsworthia and the Desulfovibrio spp., and a novel NADH-coupled, isethionate-forming sulfoacetaldehyde reductase (SarD) is found only in B. wadsworthia. The sulfite released by the GRE is reduced to sulfide by Dsr and coupled to proton translocation for ATP synthesis (symbolized as [H]) when using electrons from oxidation of an alternative electron donor, such as lactate (gray box). AckA, acetate kinase; Pta, phosphotransacetylase. (C) The gene clusters identified in this study. In B. wadsworthia, two separate clusters encode for taurine transport and conversion to isethionate (ald, tpa, and sarD), and also for isethionate desulfonation and the conversion of acetaldehyde to acetyl-CoA (adhE, islA, and islB). The Desulfovibrio strains harbor only a gene cluster for isethionate transport (dctP and fused dctMQ) and desulfonation (islA and islB), and acetaldehyde dehydrogenases are encoded elsewhere in their genomes (not depicted in C). Single-gene knockouts in D. alaskensis G20 that result in an inability to use isethionate (but not free sulfite) are indicated by squared gene symbols.