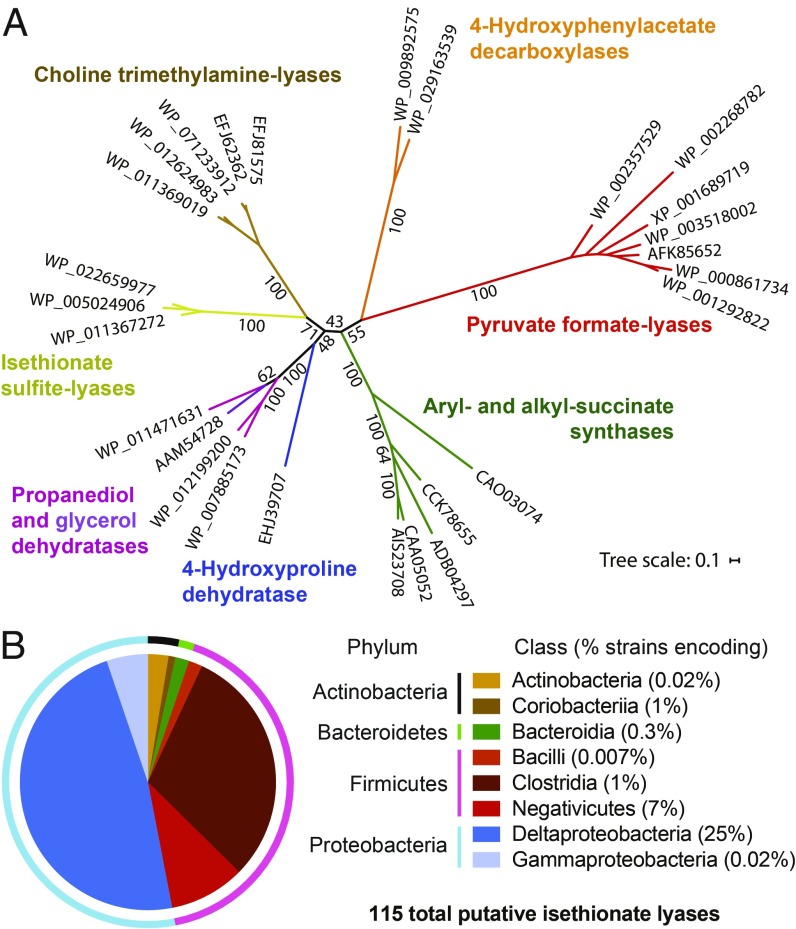
Fig. 4.
Anaerobic organosulfonate metabolism has an unexpectedly broad distribution in sequenced bacteria. (A) A maximum likelihood phylogenetic tree illustrating that the IslAs are phylogenetically distinct from previously characterized GREs. The listed sequence identifiers are GenBank accession codes. (B) Illustration of the distribution of 115 putative IslAs, as retrieved from the National Center for Biotechnology Information database at a threshold of 62% identity across sequenced bacterial genomes (SI Appendix, Table S1). The pie chart showing their relative taxonomic distribution on the phylum level (outer ring) demonstrates that the majority are found in genomes of Proteobacteria and Firmicutes, and at the class level (inner circle), that they are distributed predominantly across Deltaproteobacteria, Clostridia, and Negativicute genomes. The percentages shown in brackets indicate (at the class level) the number of genomes with putative IslAs relative to all known genome sequences for this class, confirming the enrichment of IslAs in the Deltaproteobacteria (encoded in 25% of all known genomes).