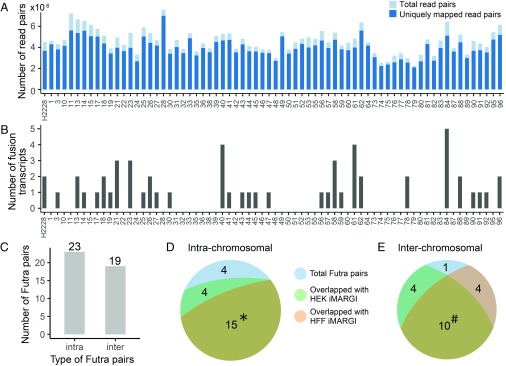
Fig. 5.
Fusion transcripts detected from the new lung cancer samples. (A) The number of RNA-seq read pairs (light blue bar) and the uniquely mapped read pairs (dark blue bar) of each sample (column). (B) Number of detected fusion transcripts (y axis) in each sample (columns). (C) Numbers of intra- and interchromosomal Futra pairs detected from the 69 cancer samples. (D and E) Intersections of these intrachromosomal (D) and interchromosomal (E) Futra pairs to RNA–DNA interactions in HEK (green) and HFF (pink) cells. : The 15 intrachromosomal Futra pairs that overlap with RNA–DNA interactions in both HEK and HFF (yellow-green) cells are ALK:EML4, RP11-557H15.4:SGK1, FRS2:NUP107, ACTN4:ERCC2, LRCH1:RP11-29G8.3, CUX1:TRRAP, CEP70:GSK3B, NIPBL:WDR70, KCTD1:SS18, CHST11:NTN4, RP1-148H17.1:TOP1, LMO7:LRCH1, NAV3:RP1-34H18.1, LPP:PPM1L, and MTOR-AS1:RERE. #: The 10 interchromosomal Futra pairs that overlap with RNA–DNA interactions in both HEK and HFF (yellow-green) cells are LIN52:PI4KA, LPP:OSBPL6, FCGBP:MT-RNR2, KMT2B:MALAT1, ETV6:TTC3, KTN1:MALAT1, FLNA:MALAT1, COL1A2:MALAT1, COL1A1:MALAT1, and MALAT1:TNFRSF10B.