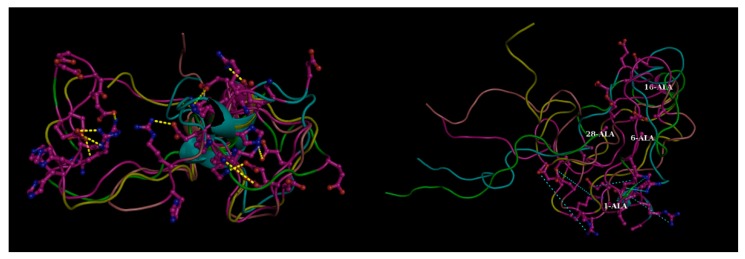
Figure 3.
Effect of mutations in disordered proteins. Four transient (flitting) salt bridge forming charged residues (1-Asp, 6-His, 16-Lys, 28-Lys) mutated to alanine in beta amyloid (Aβ42) resulting in the dismantling in salt-bridges globally throughout the structural ensemble (Left Panel: Mutant compared to the Right: Native). These transient salt-bridges continuously keep altering their partners throughout the whole simulation trajectory supporting different conformations at different time points and thereby supporting a conformational ensemble (illustrated in Figure 3. of Reference [19]). The yellow dashed lines in the left panel (native) show the salt-bridges found individually in the five randomly chosen conformers (within 4 Å) while the same connections are shown by thinner cyan dashed lines in the right panel to portray the absence of these ionic interactions (far greater than 4 Å). Molecular Dynamics simulation trajectories collected from Reference [19]. Briefly, explicit-water molecular dynamic (MD) simulation was performed with AMBER 12 at T = 300 K using the ff99SB force field with periodic boundary conditions and TIP3P water model. Figure reconstructed in Pymol.