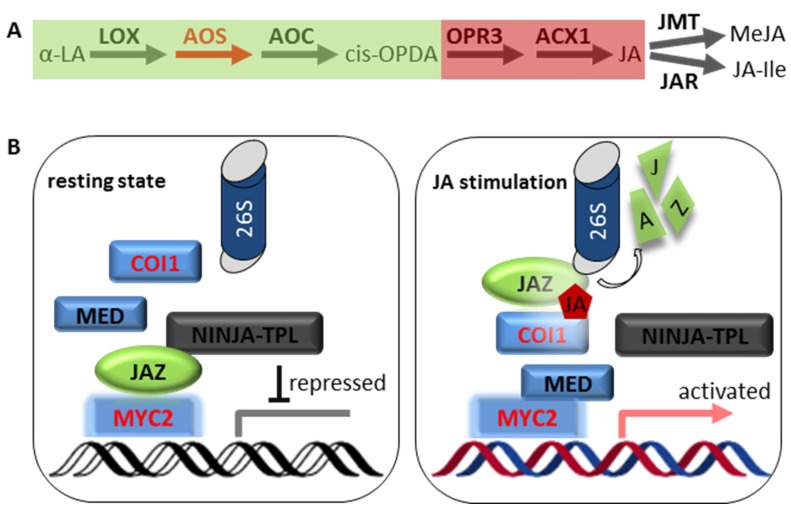
Figure 3.
Jasmonate (JA) biosynthesis and signaling transduction pathway. (A) Model for the JA biosynthesis pathway. The intermediate OPDA is synthesized in the chloroplasts. JA is synthesized in the peroxisomes and exported to the cytosol, where it is converted to other bioactive derivates (i.e., JA-Ile). The key enzyme AOS is highlighted in red. (B) Model for the JA signaling transduction pathway of the MYC-branch in Arabidopsis. In the non-induced cells (left, low JA level), MYC2 activity is repressed by JAZ proteins that interact with NINJA to recruit transcriptional repressor TPL. In the JA-stimulated cell (right, high JA level), JAZ proteins are degraded by the SCFCOI1-mediated 26S-proteosome. MYC2 is released to interact with the transcriptional mediator to activate JA-responsive gene expression. Abbreviations: α-LA, α-linolenic acid; ACX1, acyl-CoA-oxidase 1; AOC, allene oxide cyclase; AOS, allene oxide synthase; COI1, coronatine insensitive 1; JA, jasmonic acid; JA-Ile, Jasmonic acid-isoleucine conjugate; JAR, jasmonate resistant; JAZ, jasmonate ZIM domain; JMT, JA methyl transferase; LOX, 13-lipoxygenase; MeJA, methyl jasmonate; MED, mediator; NINJA, novel interactor of JAZ; OPDA, 12-oxophytodienoic; OPR3, OPDA Reductase 3; TPL, TOPLESS.